Center for Cancer Systems Therapeutics (CaST)
Research Overview
The advent of single cell approaches has dramatically improved our ability to study human disease and to appreciate its hidden complexity. This has changed the study of cancer from a largely cell-autonomous perspective, determined by individual driver mutations, to one that focuses on the dysregulation of intracellular and cell-cell interaction networks regulating the interplay between malignant cells and cells in the Tumor Microenvironment (TME). In addition to the T cells that mediated immune response, many additional TME subpopulations have emerged as playing a key role in tumor maintenance and progression, from fibroblasts (1, 2) and macrophages (3) to cellular niches contributing the paracrine signals necessary to support metastasis (4, 5). These advances are transforming cancer research from studying the average of a mixture of co-existing cell states to the characterization of each individual cell state comprising a tumor mass.
Such intra-tumor heterogeneity creates two complementary yet intimately related issues. First, virtually isogenic, yet transcriptionally distinct malignant cell states may exist within the same tumor (6, 7). These states may not only elicit distinct drug sensitivity but can also defeat monotherapy by plastically reprogramming into each other, thus first escaping targeted or immune therapy (8) and then regenerating the full heterogeneity of the tumor (6). Thus, the development of novel methodologies to inventory molecularly distinct subpopulations of transformed cells that coexist in the TME and to elucidate their genetic and pharmacological dependencies represents a critical step in the eradication of aggressive and untreatable malignancies. Second, and equally important, cancer cells leverage aberrant paracrine signals to selectively recruit (or exclude) healthy cells and/or reprogram them to create a pro-malignant TME. For instance, this is accomplished by tumor-mediated recruitment of T regulatory cells (Tregs) and other immunosuppressive subpopulations, such as macrophages (3) and Cancer Associated Fibroblasts (CAFs) (1). We thus propose that the development of methods to inventory and target pro-malignant, non-transformed TME populations is a critical step towards improving treatment of human malignancies.
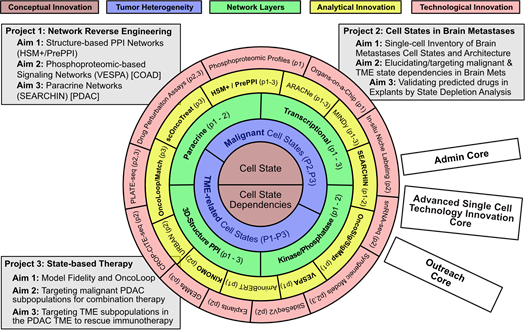
Figure 1: CaST Center at a glance. CaST is organized as a set of concentric frameworks centered around the emerging concept of Cell States and Cell State Dependencies (Layer 1, brown) and their role in defining inter-dependent malignant and TME-related subpopulations (Layer 2, purple). Elucidating the internal dependencies and interactions is predicated on the reverse engineering of Transcriptional, Signaling, 3D-Structure mediated PPI, and Paracrine networks (Layer 3, green). Reverse Engineering and Integrative Interrogation of these networks requires both analytical innovations (Layer 4, yellow, with novel algorithms shown in bold) and complementary technological innovations to (a) generate the necessary data for network reconstruction and (b) validate predictions arising from their interrogation (Layer 5, red). The Center is organized around three coherent and interdependent projects aimed at developing fully integrated computational and experimental methodologies and proof of concept (PoC) applications to address key basic-science and translational challenges in cancer. The Center is supported by three Cores that ensure proper management, access to cutting edge single cell technologies and methodology dissemination.
Rather than focusing on a specific tumor type, the proposed CaST Center is thus dedicated to developing novel network-based methodologies that integrate both computational and experimental approaches, to address these two critical challenges at the single cell level. Specifically, we will build upon technologies for the reverse engineering and interrogation of multi-layer cellular networks—comprising regulatory, signaling, protein-complex, and paracrine interactions—to produce a highly generalizable tumor- and mutation-agnostic framework for the study and treatment of human malignancies at the individual tumor cell state level (Figure 1). Thus, CaST will focus on the development of novel, interactome-based methodological frameworks that are generalizable to virtually any tumor context, as already shown in previous pancancer publications (9-12). This is consistent with a study of human malignancies that is increasingly focused on mechanism conservation and more universal dependencies, rather than on tumor organ sites. We will thus select a handful of tumor contexts as Proof-of-Concept (PoC) for different methodologies, based on (a) availability of critical reagents and datasets to successfully achieve PoC and (b) tumor biology expertise and track record of CaST investigators. Specifically:
- In Project 1, which focuses on the reverse engineering of multi-layer interactomes, we will study colon adenocarcinoma (COAD) due to availability of a unique, large-scale dataset of time-dependent phosphoproteomic profiles generated following drug perturbations and pancreatic ductal adenocarcinoma (PDAC) because it represents the archetype of stroma-rich tumors and is thus ideally suited to elucidate paracrine interactions between malignant and TME cells. LEARN MORE ABOUT PROJECT 1
- In Project 2, we will exclusively focus on brain metastases from the two malignancies that most contribute to this phenotype, melanoma (SKCM) and non-small-cell lung cancer (NSCLC). LEARN MORE ABOUT PROJECT 2
- Finally, in Project 3, we will leverage an extensive prostate carcinoma (PCa) GEMM resource to investigate the concept of cancer model fidelity and will extend our PDAC studies to predict and validate therapies aimed at depleting individual malignant and immunosuppressive cell states for combination therapy. LEARN MORE ABOUT PROJECT 3
All CaST-developed methodologies will be fully generalizable and adaptable to study other tumors with minimal parameter tuning. We believe that such an approach is most responsive to the CSBC mission and will provide tools that other consortium centers, and the broader cancer research community at large, will be able to leverage for their studies.
References
1. Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, Teinor JA, Belleau P, Biffi G, Lucito MS, Sivajothi S, Armstrong TD, Engle DD, Yu KH, Hao Y, Wolfgang CL, Park Y, Preall J, Jaffee EM, Califano A, Robson P, Tuveson DA. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer discovery. 2019;9(8):1102-23. Epub 2019/06/15. doi: 10.1158/2159-8290.CD-19-0094. PubMed PMID: 31197017.
2. Affo S, Nair A, Brundu F, Ravichandra A, Bhattacharjee S, Matsuda M, Chin L, Filliol A, Wen W, Song X, Decker A, Worley J, Caviglia JM, Yu L, Yin D, Saito Y, Savage T, Wells RG, Mack M, Zender L, Arpaia N, Remotti HE, Rabadan R, Sims P, Leblond AL, Weber A, Riener MO, Stockwell BR, Gaublomme J, Llovet JM, Kalluri R, Michalopoulos GK, Seki E, Sia D, Chen X, Califano A, Schwabe RF. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell. 2021;39(6):883. Epub 2021/06/16. doi: 10.1016/j.ccell.2021.05.010. PubMed PMID: 34129825.
3. Obradovic A, Chowdhury N, Haake SM, Ager C, Wang V, Vlahos L, Guo XV, Aggen DH, Rathmell WK, Jonasch E, Johnson JE, Roth M, Beckermann KE, Rini BI, McKiernan J, Califano A, Drake CG. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell. 2021;184(11):2988-3005 e16. Epub 2021/05/22. doi: 10.1016/j.cell.2021.04.038. PubMed PMID: 34019793; PMCID: PMC8479759.
4. Lee JW, Stone ML, Porrett PM, Thomas SK, Komar CA, Li JH, Delman D, Graham K, Gladney WL, Hua X, Black TA, Chien AL, Majmundar KS, Thompson JC, Yee SS, O'Hara MH, Aggarwal C, Xin D, Shaked A, Gao M, Liu D, Borad MJ, Ramanathan RK, Carpenter EL, Ji A, de Beer MC, de Beer FC, Webb NR, Beatty GL. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature. 2019;567(7747):249-52. Epub 2019/03/08. doi: 10.1038/s41586-019-1004-y. PubMed PMID: 30842658; PMCID: PMC6430113.
5. Massague J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529(7586):298-306. Epub 2016/01/23. doi: 10.1038/nature17038. PubMed PMID: 26791720; PMCID: PMC5029466.
6. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, Gritsch S, Perez EM, Gonzalez Castro LN, Lan X, Druck N, Rodman C, Dionne D, Kaplan A, Bertalan MS, Small J, Pelton K, Becker S, Bonal D, Nguyen QD, Servis RL, Fung JM, Mylvaganam R, Mayr L, Gojo J, Haberler C, Geyeregger R, Czech T, Slavc I, Nahed BV, Curry WT, Carter BS, Wakimoto H, Brastianos PK, Batchelor TT, Stemmer-Rachamimov A, Martinez-Lage M, Frosch MP, Stamenkovic I, Riggi N, Rheinbay E, Monje M, Rozenblatt-Rosen O, Cahill DP, Patel AP, Hunter T, Verma IM, Ligon KL, Louis DN, Regev A, Bernstein BE, Tirosh I, Suva ML. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell. 2019;178(4):835-49 e21. Epub 2019/07/23. doi: 10.1016/j.cell.2019.06.024. PubMed PMID: 31327527.
7. Yuan J, Levitin HM, Frattini V, Bush EC, Boyett DM, Samanamud J, Ceccarelli M, Dovas A, Zanazzi G, Canoll P, Bruce JN, Lasorella A, Iavarone A, Sims PA. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018;10(1):57. Epub 2018/07/26. doi: 10.1186/s13073-018-0567-9. PubMed PMID: 30041684; PMCID: PMC6058390.
8. Risom T, Langer EM, Chapman MP, Rantala J, Fields AJ, Boniface C, Alvarez MJ, Kendsersky ND, Pelz CR, Johnson-Camacho K, Dobrolecki LE, Chin K, Aswani AJ, Wang NJ, Califano A, Lewis MT, Tomlin CJ, Spellman PT, Adey A, Gray JW, Sears RC. Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nature communications. 2018;9(1):3815. Epub 2018/09/21. doi: 10.1038/s41467-018-05729-w. PubMed PMID: 30232459; PMCID: PMC6145927.
9. Paull EO, Aytes A, Jones SJ, Subramaniam PS, Giorgi FM, Douglass EF, Tagore S, Chu B, Vasciaveo A, Zheng S, Verhaak R, Abate-Shen C, Alvarez MJ, Califano A. A modular master regulator landscape controls cancer transcriptional identity. Cell. 2021;184(2):334-51 e20. Epub 2021/01/13. doi: 10.1016/j.cell.2020.11.045. PubMed PMID: 33434495.
10. Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CE, Cancer Genome Atlas Research N, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich L. The Immune Landscape of Cancer. Immunity. 2018;48(4):812-30 e14. Epub 2018/04/10. doi: 10.1016/j.immuni.2018.03.023. PubMed PMID: 29628290; PMCID: PMC5982584.
11. Broyde J, Simpson DR, Murray D, Paull EO, Chu BW, Tagore S, Jones SJ, Griffin AT, Giorgi FM, Lachmann A, Jackson P, Sweet-Cordero EA, Honig B, Califano A. Oncoprotein-specific molecular interaction maps (SigMaps) for cancer network analyses. Nat Biotechnol. 2021;39(2):215-24. Epub 2020/09/16. doi: 10.1038/s41587-020-0652-7. PubMed PMID: 32929263; PMCID: PMC7878435.
12. Mundi PS, Dela Cruz FS, Grunn A, Diolaiti D, Mauguen A, Siddiquee A, You D, Realubit R, Karan C, Ortiz MV, Accordino M, Aloni Z, Brogan F, Bruce J, Caescu CI, Carvajal R, Crew K, Decastro G, Heaney M, Henick B, Hershman D, Hou J, Iwamoto F, Jurcic J, Kiran PR, Kluger M, Kreisl T, Lamanna N, Lassman A, Lim E, Manji G, McKhann G, McKiernan J, Neugut A, Olive K, Rosenblat T, Schwartz GK, Shu C, Sisti M, Tergas A, Vattakalam R, Welch M, Wenske S, Wright J, Hibshoosh H, Kalinsky K, Aburi M, Sims PA, Alvarez M, Kung AL, Califano A. Pre-clinical validation of an RNA-based precision oncology platform for patient-therapy alignment in a diverse set of human malignancies resistant to standard treatments. bioRxiv 20211003462951. 2021.
13. Lo YH, Kolahi KS, Du Y, Chang CY, Krokhotin A, Nair A, Sobba WD, Karlsson K, Jones SJ, Longacre TA, Mah AT, Tercan B, Sockell A, Xu H, Seoane JA, Chen J, Shmulevich I, Weissman JS, Curtis C, Califano A, Fu H, Crabtree GR, Kuo CJ. A CRISPR/Cas9-engineered ARID1A-deficient human gastric cancer organoid model reveals essential and non-essential modes of oncogenic transformation. Cancer discovery. 2021. Epub 2021/01/17. doi: 10.1158/2159-8290.CD-20-1109. PubMed PMID: 33451982.
14. Affo S, Nair A, Brundu F, Ravichandra A, Bhattacharjee S, Matsuda M, Chin L, Filliol A, Wen W, Song X, Decker A, Worley J, Caviglia JM, Yu L, Yin D, Saito Y, Savage T, Wells RG, Mack M, Zender L, Arpaia N, Remotti HE, Rabadan R, Sims P, Leblond AL, Weber A, Riener MO, Stockwell BR, Gaublomme J, Llovet JM, Kalluri R, Michalopoulos GK, Seki E, Sia D, Chen X, Califano A, Schwabe RF. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell. 2021;39(6):866-82 e11. Epub 2021/05/01. doi: 10.1016/j.ccell.2021.03.012. PubMed PMID: 33930309; PMCID: PMC8241235.