News
Uniting Structural and Systems Biology: An Interview with Barry Honig

When Columbia University founded the Center for Multiscale Analysis of Genomic and Cellular Networks (MAGNet) in 2005, one of its goals was to integrate the methods of structural biology with those of systems biology. Considering protein structure within the context of computational models of cellular networks, researchers hoped, would not only improve the predictive value of their models by giving another layer of evidence, but also lead to new types of predictions that could not be made using other methods.
In a new paper published in Nature magazine, Barry Honig, Andrea Califano, and other members of the Columbia Initiative in Systems Biology, including first authors Qiangfeng Cliff Zhang and Donald Petrey, report that this goal has now been realized. For the first time, the researchers have shown that information about protein structure can be used to make predictions about protein-protein interactions on a genome-wide scale. Their approach capitalizes on innovative techniques in computational structural biology that the Honig lab has developed over the last 15 years, culminating in the development of a new algorithm called Predicting Protein-Protein Interactions (PrePPI). In this interview, Honig describes the evolution of this new approach, and what it could mean for the future of systems biology.
Barry Honig is Professor of Biochemistry and Molecular Biophysics, director of the Center for Computational Biology and Bioinformatics, and an investigator of the Howard Hughes Medical Institute.
Why hasn’t information about protein structure been incorporated into systems biology in the past?
The impact of structural biology has been enormous, though it has tended to focus on describing biology at a very small scale. Over time, scientists have built a repository called the Protein Data Bank (PDB), which contains over 70,000 structures of proteins. Historically, being able to represent the structure of a protein in atomic detail has made it possible to understand its specific biological function very well.
Although the PDB contains a significant number of proteins and covers many organisms, structural information has not kept up with the explosive growth of sequence information. In addition, there are far fewer structures for protein-protein complexes than for individual proteins. So on a systems biology scale, where we want to achieve a genome-wide perspective on how proteins interact, we haven’t in the past been able to use three-dimensional structural information.
In recent years, you’ve developed an unconventional approach to detecting relationships between proteins. Could you describe this?
When you look closely at the PDB, it turns out that many proteins are very similar in shape. If you limit yourself to proteins that are fundamentally different, there are maybe 25,000 proteins. As the database has grown, there has been a lot of thinking about what people call protein structure space. That is, how many truly different proteins are there across all organisms? People have attempted to classify proteins into different groups. Some of the classifications are based on their function — kinases, for example — and others are based on structural similarity.
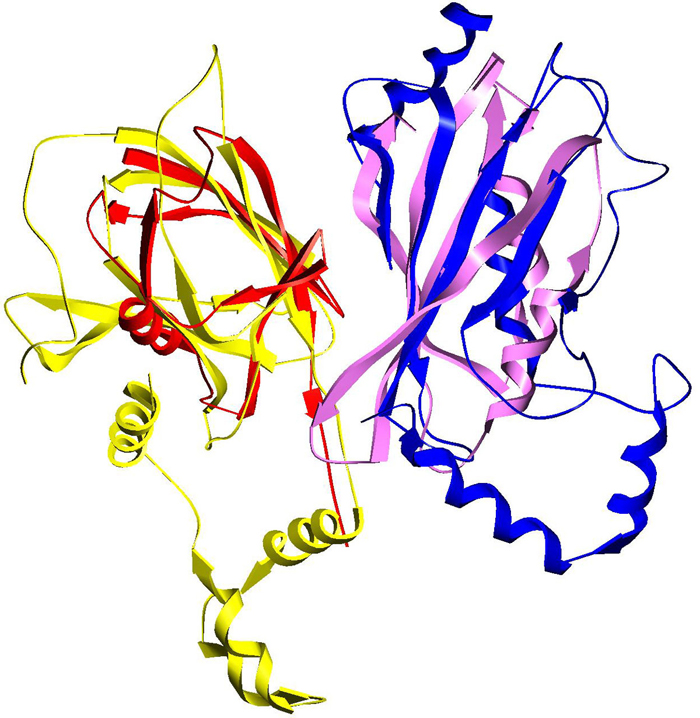
An example of PrePPI interaction predictions using remote structural homology. A template complex involving two ubiquitination pathway proteins (red and blue) is used to predict an interaction between EF1-delta and VHL (yellow and purple).
It is widely known that protein structure is better conserved than protein sequence. If a molecular biologist finds a sequence of a new protein, he or she might run a program like BLAST, which looks through millions of sequences to find a protein with a similar sequence. The problem is that many proteins look alike physically in space — and therefore might be expected to function in similar ways — but because their sequences are different these relationships can’t be detected by BLAST. The minute that two sequences are so different that they are statistically unrelated, you lose information that could establish a relationship. Structural biology can find those relationships. Using an approach called structural alignment, proteins have been grouped based on their overall folds, the shape of a protein that a sequence of amino acids folds into.
What I and others in my lab have been saying for years, however, is that even this approach to classification loses information. There are many proteins that have very similar structures in certain regions but are different everywhere else. Just as sequence searches look for local regions within a sequence where amino acids are similar, we have argued that we should be looking for local regions where proteins have similar structures, even if, globally, the proteins look quite different. Over the years we have shown that proteins can have local regions with different sequence but similar structure and similar function.
What does this expansion of possible relationships between proteins mean in terms of how you detect protein-protein interactions?
Our driving hypothesis has been that most interactions between any two proteins bear some similarity to one of the protein complexes that have been catalogued in the PDB. To predict how those proteins interact, we just need to find the right complex in the PDB. And so in our new paper in Nature , we describe methods for looking broadly to find relationships between proteins that could possibly lead to a prediction of a protein-protein interaction.
Say that I want to know whether two proteins — protein A and protein B — interact. For protein A, I can search for proteins with similar structures. I might, for example, find 1,000 proteins that have a similar structure in a local region. If I look for protein B and its neighbors — proteins with some structural similarity — I might also find 1,000 of those. If I then cross these two collections, there are one million possible pairs. If any one of those pairs is known to form a protein complex, then I’ve established a relationship between my two starting proteins A and B. By using all of protein structure space as a source of information, we can now predict the probability that those original two proteins interact, possibly in similar ways. By looking at relationships between proteins that can only be identified by shape, this approach greatly expands the value of structural information. Without taking this step the number of relationships that can be detected is significantly reduced.
Earlier you said that the Protein Data Bank contains relatively few experimentally confirmed protein structures. Wouldn’t that limit the usefulness of this approach?
Your question touches on what’s great about computational modeling. If you know a sequence of amino acids in a protein, you can search the PDB for similar sequences where the structure is known. We know that specific sequences produce specific structures, and so identifying these similarities in sequence lets you develop a structural model of the protein whose structure is not known. This procedure is known as homology modeling. Our new algorithm, called PrePPI (Predicting Protein-Protein Interactions), incorporates these homology models, and has helped us to identify 300,000 high-probability interactions in humans. If we had only used structural information from experimentally determined models in the PDB we would find only about 30,000. The difference between the two numbers is a dramatic demonstration of the impact of homology modeling.
This approach is new — both for structural and systems biology. Historically, structural biologists, including myself, have focused on looking at protein structures in great detail. What we are now saying for the purposes of systems biology is that we are using structure to give us statistical clues, and we don’t need our structures to be perfect to get that clue. By lowering our criteria and going a little fuzzy, we can expand the usefulness of structural information to generate predictive models of protein-protein interactions.
How do you determine the likelihood that a predicted protein-protein interaction is real?
Another critical element of this approach is that we use Bayesian statistics to evaluate each predicted relationship and generate a probability that it is meaningful. We don’t say that one interaction is real and another is not. We identify relationships within our comparative structural approach and then assign a value to each one.
A high predictive value gives us one piece of information, but if there is another independent clue that indicates, for example, that the two proteins are co-expressed or that they have the same phylogenetic history, Bayesian statistics allows us to multiply the individual likelihoods. Integrating totally different sources of information — structure and co-expression — allows us to pull signals out of noisy structural information. Protein structure has not been used in this way before.
Why might this approach be more useful than other methods for detecting protein-protein interactions?
Biologists from many fields are searching for hypotheses for what proteins interact with one another. Until now, they have primarily relied on experimental methods such as yeast two-hybrid, which can generate predictions but is often unreliable because it cannot duplicate biological conditions. Our approach replaces an uncertain experimental method with an uncertain computational method, but has the advantage that all you have to do is access the PrePPI website; with a few clicks of a mouse you can obtain the probability that a particular interaction is real. And we think that many of the predictions are real. Moreover, in most cases we are able to provide a structural model for the interaction.
Once you have a prediction and a structural model you can then test the model experimentally. You could design an experiment in which you mutate a protein precisely where PrePPI says that it interacts, giving you a level of precision in testing hypotheses that hasn’t been possible before. It takes the huge body of structural information we’ve generated over the last 30-40 years and uses it in new ways. As the paper shows, structural information turns out to be more valuable than other sources of information, because it gives you a physical model of what’s happening. It takes you a little further out of the fog than other sources of information.
In the paper you describe several examples of ways in which you have applied PrePPI to generate new biological discoveries. Can you talk about one?
One area of biology in which I’m particularly interested is a class of proteins called protocadherins. Tom Maniatis, one of our collaborators, showed that these proteins are involved in neuronal self-avoidance. Protocadherins somehow accomplish this function but no one knows how. Using PrePPI, we predicted that a protocadherin interacts with a tyrosine kinase receptor and then verified it experimentally in the lab. This is a very preliminary finding, but PrePPI has now provided us with a new direction to help us understand the molecular basis of protocadherin function.
What’s next for PrePPI?
Right now we have a yeast database and a human database, but we want to create new databases for other organisms and develop the ability to compare interactions across organisms. For example, we’re beginning to work with Raul Rabadan and Sagi Shapira of the Columbia Initiative in Systems Biology on host-pathogen interactions. Another potential application is in the study of cancer pathways. More generally, we’re hoping to apply it broadly with collaborators. I’m really looking forward to being involved in some interesting applications.
We’re also planning to improve the methods in PrePPI itself, introducing new information and dealing with types of interactions that aren’t included. Everything in PrePPI now involves two proteins with structures that interact. There are many interactions where one protein has structure and the other is an unstructured peptide. Many protein-protein interactions are of that kind and we’re beginning to think of ways to deal with them. Being able to do so would vastly expand PrePPI’s reach.
— Interview by Chris Williams
Related publication
Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012 Oct 25;490(7421):556-60.