News
Automated, Single Cell RNA-Seq Enables Research on Tumor Ecology
Assistant Professor Peter Sims and postdoctoral research scientist Jinzhou Yuan displaying their platform for automated single cell RNA-Seq. Photo: Lynn Saville.
RNA sequencing (RNA-Seq) has become a workhorse technology for research in systems biology. Unlike genome sequencing, which reveals a sample’s DNA blueprint, RNA-Seq catalogs the constantly changing transcriptome; that is, it itemizes and quantifies the complete set of messenger RNA transcripts that are present in cells at a specific time and under specific conditions. In this way, RNA-Seq makes it possible to investigate how the information encoded in the genome is functionally transformed into observable traits, and provides valuable data for defining and comparing different biological states.
Conventional RNA-Seq generates an average summary of mRNA abundance across all of the cells in a sample. Recent research, however, has created a demand for higher resolution technologies capable of generating mRNA profiles at the level of single cells. In cancer biology, for example, there is an increasingly acute awareness that gene expression in the cells that make up malignant tumors is highly heterogeneous. This suggests that in order to understand how the cells work together to drive a tumor’s cancerous behavior, scientists need better methods for characterizing the entire ecology of cells of which it is made. Being able to quantify differences in gene expression cell by cell could be one valuable way to explore such complex environments and understand how they sustain malignancy.
Although several single cell RNA-seq technologies have been unveiled in the past two years, they are expensive to operate and are not optimized to produce data on the scale that is required for systems biology research, particularly in tissue specimens with limited numbers of cells. In a new paper just published in the journal Scientific Reports, however, researchers in the laboratory of Department of Systems Biology Assistant Professor Peter Sims describe a novel approach that offers several important advantages over other existing methods.
The new, automated platform builds on previous innovations in the Sims Lab to offer a cheap, efficient, and reliable way to simultaneously measure gene expression in thousands of individual cells from a single tissue sample. Using custom designed microwell plates, microfluidics, temperature control systems, and software, the technology captures, tags, and generates a readout of the complete transcriptome in each cell, providing robust data that can then be analyzed to distinguish functional diversity among the cells in the sample. Already, the technology is playing a key role in several research projects being conducted in the Department of Systems Biology and promises to become even more powerful as the field of single cell genomics continues to evolve.
How the technology works
The foundation of the Sims Lab’s approach is to use custom plates made from PDMS, a type of silicone rubber. Plates can contain arrays of up to 150,000 tiny wells, each with a volume of just 100 picoliters. After individual cells from a tissue sample have been separated and suspended in solution, the microfluidic device passes the suspension over the plates and individual cells settle into single wells due to gravity. Each well is then loaded with a polymer bead that is covered with millions of copies of an oligonucleotide that has a bead-specific nucleotide sequence.
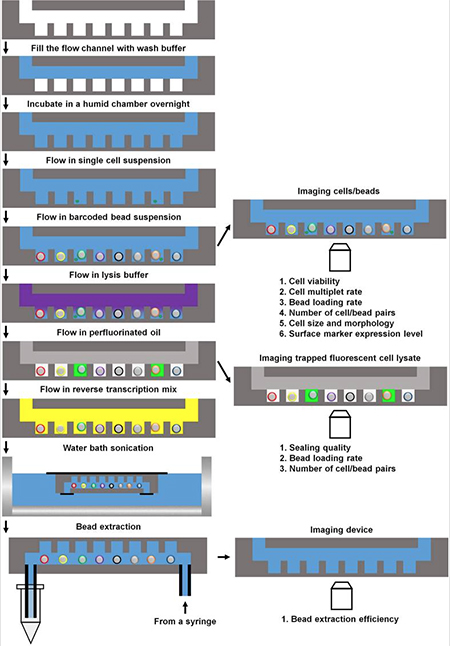
A schematic overview of the Sims Lab's automated single cell RNA-Seq platform. After single cells settle from solution into individual wells, barcoded beads are inserted, the cells go through lysis, and the microwells are sealed with oil. The mRNA from each cell goes through reverse transcription and the resulting cDNA is tagged with the oligonucleotide barcodes. During the experiment, the plate can be removed to observe things such as cell size and shape, expression of fluorescent markers, and the health of the cells about to be sequenced. Courtesy Scientific Reports.
Once the beads are loaded in the wells, the cells are exposed to a chemical that causes them to burst, and the entire microwell array is sealed with a thin layer of oil to prevent cross-contamination among the wells. The released mRNA molecules are then captured by the oligonucleotides and exposed to another reaction that causes reverse transcription. The resulting complementary DNAs (cDNAs) end up labelled with the bead-specific nucleotide sequence and stick to the bead. Because the oligonucleotide sequence is unique to a single cell in a single well, it serves as a “barcode” that can later be used to identify the specific cell from which a particular cDNA came.
The benefit of this barcode is that rather than sequencing the cells one at a time, all of the beads can then be removed from the microwell plate and mixed together in one tube. They are subjected to another reaction that separates the oligonucleotide–cDNA molecules from the beads and then the entire pool of cDNA is sequenced at the same time at the Columbia Genome Center. When run through a standard Illumina sequencer, the scientists then generate a readout in which each molecule’s sequence contains its barcode. In aggregate this reveals the distinctive transcriptome for each of the single captured cells.
This general approach, initially implemented by Sayantan Bose, a former postdoctoral research scientist in the Sims Lab, was first published in 2015. Since that time, the lab welcomed Jinzhou Yuan, a postdoc whose background is in engineering and who developed a method to automate the workflow. The new platform just reported in Scientific Reports makes the process fast, easy to use, and capable of efficiently preparing cells for RNA sequencing in a high throughput manner.
Unique benefits and opportunities
The Sims Lab’s approach offers a number of important advantages over other existing technologies. For one, the pooled approach to sequencing cDNA from thousands of cells simultaneously, as well as the bespoke nature of the tool, mean that it is much less expensive to operate than other more widely used commercial platforms. The new high throughput microfluidics setup makes it possible to prepare single cell RNA-Seq libraries for just 10 cents per cell. This is a tremendous advantage for generating statistically robust single cell data from tissue samples with high cellular heterogeneity.
The new automation has also led to a marked improvement in the efficiency with which the scientists can capture cells from precious tissue samples. The latest paper reports that more than 50% of the cells that are initially loaded into the microfluidic device can be captured by microwells. “We set out to make the platform easier to use,” Sims says, “but as often happens when you implement automation we also got a dramatic increase in performance. The enhanced capability of the technology means that it produces much better data.”
A final important advantage of the Sims Lab’s platform derives from its use of solid state microwell plates for cell capture. Other competing experimental technologies have tried to solve the problem of isolating single cells by encasing them in oil droplets. Those approaches, however, do not allow an investigator to image cells in the device for quality control before sending them for sequencing.
"The future of single cell genomics lies in integrating multiple observables... We think we should be well positioned to participate in where the field is going."
At various stages in the Sims Lab’s workflow it is possible to remove a plate and look at it under a microscope, offering an opportunity to confirm, for example, that multiple cells aren’t stuck together in a well and that the cells that are about to be sequenced are healthy. Microwell arrays also make it possible to perform additional experiments on the same cells in parallel, such as staining them with fluorescent probes to gather phenotypic information. This opens the door to conducting more complex, integrative experiments that could lead to subtler insights into the diversity of the cells being studied.
Sims explains that this capability is essential for some of the research being planned in the Department of Systems Biology. “It’s becoming increasingly clear,” Sims explains, “that the future of single cell genomics lies in integrating multiple observables in addition to mRNA. Because we are able to isolate and do things to cells in microwells during our experiments, we think we should be well positioned to participate in where the field is going.”
Dissecting brain tumors, one cell at a time
Already the Sims Lab’s single cell RNA-seq technology has been attracting interest from other scientists in the Department of Systems Biology and other corners of Columbia University Medical Center (CUMC). As a participating investigator in the Center for Topology of Cancer Evolution and Heterogeneity, a center in the National Cancer Institute’s Physical Sciences-Oncology Network (PSON), Sims is an essential player in a project focused on dissecting cellular heterogeneity in glioblastoma multiforme (GBM), the most common and deadly form of brain cancer.
The project relies on collaboration within a remarkable interdisciplinary team, beginning when neurosurgeon Jeff Bruce gathers GBM tissue during brain surgery. He sends these samples to Peter Canoll, Director of Neuropathology at CUMC, who categorizes the tissue using standard pathology methods. In turn, Canoll then sends them to Anna Lasorella, an associate professor in the Institute for Cancer Genetics, whose lab has expertise in safely dissociating a tumor into single cells. It is at this point that Jinzhou Yuan in the Sims Lab receives the sample, which is then passed through the automated sequencing workflow.
The endpoint of the process comes when the single cell RNA-Seq data become available for analysis. Leading this effort is Raul Rabadan, a theoretical physicist in the Department of Systems Biology, who uses concepts from a mathematical field called topological data analysis to categorize the individual cells that have gone through the sequencing pipeline. His approaches are revealing the composition of brain tumors in a much more nuanced way than has previously been possible.
"The single cell platforms Peter Sims has been pioneering at Columbia are allowing us to study some important biological problems," Rabadan says. "This includes heterogeneity among the cells that make up cancer."
The scalable, high throughput, and efficient nature of the technology is a key factor in moving toward this scientific goal. “Often,” Sims explains, “cancer researchers just grind up a tumor and sequence the whole thing. A few years ago it also became possible to select specific cells and sequence a small number of them. Now, our single cell RNA-Seq platform gives us a high enough throughput that we can simply sequence all of the cells, providing an unbiased picture of the entire tumor environment. This gives us a very clear, more objective view of what’s actually there without having to try to guess which markers we should be tracking.”
Although research on this glioblastoma project only started in the summer of 2016, it is already clear that gene expression diversity is very rich in solid tumors, a fact that will affect how we understand and potentially treat cancer in the future.
A core component in cancer systems biology research
The Sims Lab is also playing an important role in the recently opened Center for Cancer Systems Therapeutics (CaST), one of four inaugural centers in the National Cancer Institute’s new Cancer Systems Biology Consortium. In this case, Sims is not an investigator on a specific project, but directs its Molecular Profiling Core, which will support a variety of research by CaST scientists. In addition to single cell RNA-Seq, the Core is also providing a platform that integrates genome wide expression profiling with high throughput screening, as well as microfluidics and other high throughput single cell sequencing methods.
In one project that CaST plans to pursue, for example, Department of Systems Biology Chair Andrea Califano will for the first time apply algorithms his laboratory has developed — which use gene expression data to infer regulatory network models — to the study of single cells. In another, Raul Rabadan will be investigating how the cellular makeup of a tumor evolves over time in response to drug therapies and other environmental perturbations, an issue that is important in the development of drug resistance.
“Systems biology is a technology driven field, and it’s very collaborative,” Sims observes. “Usually large centers like CaST are built around a couple of core scientific concepts and a couple of technologies. I’m very happy that my lab has been able to contribute technologies that are making it possible to do such exciting science.”
— Chris Williams
Related publications
Yuan J, Sims PA. An automated microwell platform for large-scale single cell RNA-Seq. Sci Rep. 2016 Sep 27;6:33883.
Bose S, Wan Z, Carr A, Rizvi AH, Vieira G, Pe'er D, Sims PA. Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biol. 2015 Jun 6;16:120.