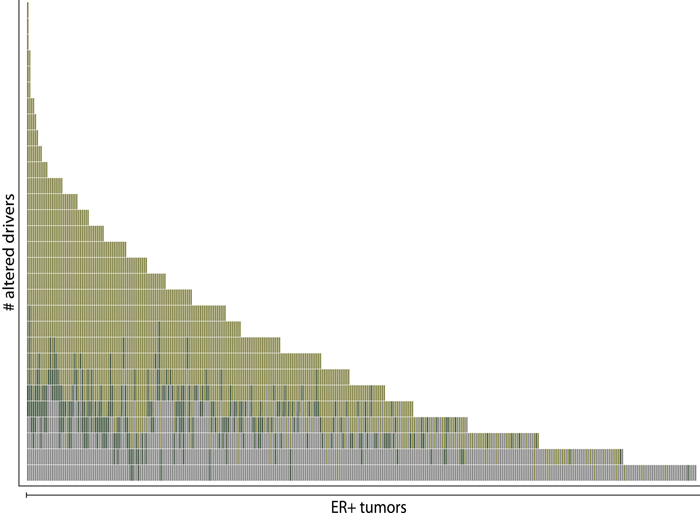
In comparison with a previous study (Stephens et al., 2012, shown in gray), a new computational approach that focuses on somatic copy number mutations increased the number of known driver mutations in breast tumors to a median of five for each tumor. The findings could raise the likelihood of finding actionable targets in individual patients with breast cancer.
For many years, researchers have known that somatic copy number alterations (SCNA’s) — insertions, deletions, duplications, and transpositions of sections of DNA that are not inherited but occur after birth — play important roles in causing many types of cancer. Indeed, most recurrent drivers of epithelial tumors are copy number alterations, with some found in up to 40% of patients with specific tumor types. However, because SCNA’s occur when entire sections of chromosomes become damaged, biologists have had difficulty developing effective methods for distinguishing genes within SCNA’s that actually drive cancer from those genes that might lie near a driver but do not themselves cause disease.
Helios nearly doubled the number of high-confidence predictions of breast cancer drivers.
In a new paper published in Cell, researchers in the laboratories of Dana Pe’er (Columbia University Departments of Systems Biology and Biological Sciences) and Jose Silva (Icahn School of Medicine at Mount Sinai) report on a new computational algorithm that promises to dramatically improve researchers’ ability to identify cancer-driving genes within potentially large SCNA’s. The algorithm, called Helios, was used to analyze a combination of genomic data and information generated by functional RNAi screens, enabling them to predict several dozen new SCNA drivers of breast cancer. In follow-up in vitro experimental studies, they tested 12 of these predictions, 10 of which were validated in the laboratory. Their findings nearly double the number of breast cancer drivers, providing many new opportunities towards personalized treatments for breast cancer. Their methodology is general and could also be used to locate disease-causing SCNA’s in other cancer types.
Leading this effort was Felix Sanchez-Garcia, a recent PhD graduate from the Pe’er Lab and a first author on the paper. The story of how this breakthrough came about illuminates how the interdisciplinary research and education that take place at the Department of Systems Biology can address important challenges facing biological and biomedical research.