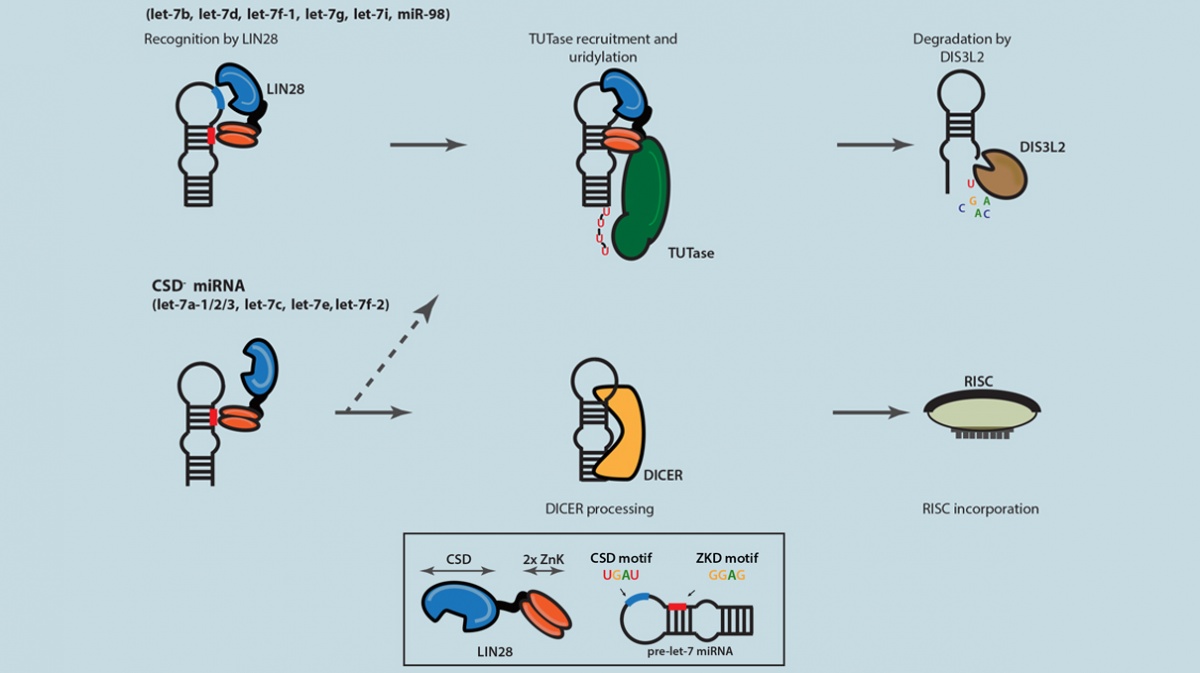
The proposed model of selective Let-7 microRNA suppression modulated by the bipartite LIN28 binding.(Image courtesy of Zhang Lab)
A new study led by Chaolin Zhang, PhD , assistant professor of systems biology , published today as the cover story of Molecular Cell , sheds light on a critical RNA-binding protein that is widely researched for its role in stem cell biology and its ties to cancer progression in multiple tissues.
The LIN28 RNA-binding protein, initially found in worms about 15 years ago, is specifically expressed in stem cells. It became well known because the protein is one of the four factors that were used to “reprogram” skin cells to induced pluripotent stem cells, or iPSCs, a breakthrough that was awarded the Nobel Prize in 2012. More recently, it was determined that the LIN28 RNA-binding protein can also be reactivated in cancer to drive tumor growth and progression. Due to its critical importance in developmental and cancer biology, scientists want to understand the role LIN28 plays at the molecular level. This new study provides some understanding of how the LIN28 protein suppresses a specific family of microRNAs, called Let-7, which are selectively lost in cancer.
“Let-7 microRNAs are the major downstream targets controlled by LIN28 identified so far. While LIN28 is mostly found in stem cells, Let-7 is only detected in differentiated cells because of stem cell-specific suppression by LIN28. However, the interplay between the two is still not well understood,” says Dr. Zhang, who is also a member of Columbia University’s Center for Motor Neuron Biology and Disease . “This study contributes to our understanding of how LIN28 suppresses Let-7, as well as provides a refined model for this important, rather complex molecular pathway.”