News
A Topological Approach to Modeling Evolution
 In Charles Darwin's seminal treatise On the Origin of Species there is only one image, which visualizes evolution as following a branching pattern in which species diverge into lineages over time like the limbs on a tree. With the increasing availability of genomic data, scientists have attempted to understand evolution at the molecular level by using a similar phylogenetic paradigm, but as Department of Systems Biology Assistant Professor Raul Rabadan , MD/PhD student Joseph Chan, and Stanford University mathematician Gunnar Carlsson point out in a new paper published in the Proceedings of the National Academy of Sciences , it has a number of shortcomings when applied in this way. By developing a new mathematical approach based on a method called persistent homology, the researchers produced several insights into viral evolution that could not be found using other means.
In Charles Darwin's seminal treatise On the Origin of Species there is only one image, which visualizes evolution as following a branching pattern in which species diverge into lineages over time like the limbs on a tree. With the increasing availability of genomic data, scientists have attempted to understand evolution at the molecular level by using a similar phylogenetic paradigm, but as Department of Systems Biology Assistant Professor Raul Rabadan , MD/PhD student Joseph Chan, and Stanford University mathematician Gunnar Carlsson point out in a new paper published in the Proceedings of the National Academy of Sciences , it has a number of shortcomings when applied in this way. By developing a new mathematical approach based on a method called persistent homology, the researchers produced several insights into viral evolution that could not be found using other means.
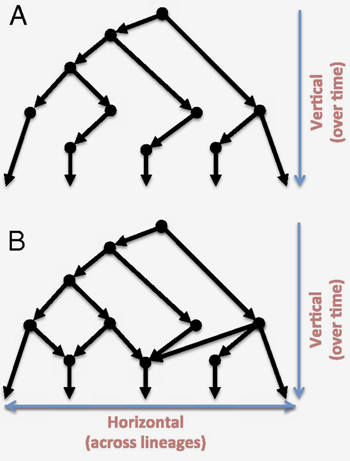
Recent genomic studies have made it clear that evolution does not only proceed in a "vertical" pattern in which one organism inherits genomic information from the organisms from which it descends (figure A). Scientists now understand that genomic evolution can also be "horizontal"; that is, genomic information can be transferred between organisms or evolutionarily similar groups of organisms that are in parallel lineages (figure B), such as in cases of species hybridization in eukaryotes, lateral gene transfer in bacteria, recombination and reassortment in viruses, viral integration in eukaryotes, and fusion of genomes of symbiotic species. These observations suggest that phylogenetic trees have limitations in their ability to characterize evolution at the molecular level and that another model is needed that can integrate both vertical and horizontal evolution.
Dr. Rabadan lectures on the topology of viral evolution at a Workshop on Topology in the School of Mathematics at the Institute for Advanced Study in Princeton, NJ. Click image to see video.
Before becoming involved in computational and systems biology years ago as a postdoctoral researcher at the Institute for Advanced Study, Dr. Rabadan was trained as a theoretical physicist, and so turned to a mathematical field called algebraic topology for a new approach to this problem. Using a method called persistent homology, the researchers analyze viral and synthetic genomic data and represent evolutionary relationships as higher-dimensional objects. Their approach uses a computer algorithm to produce a topological graph that can then be investigated visually to identify interesting evolutionary patterns.
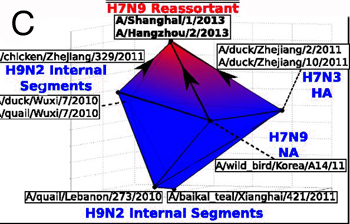
To test their method, the investigators applied it to genomic data from several viruses, including dengue virus, HIV, and influenza. They were able to detect both vertical and horizontal forms of evolution, determine the rate of horizontal genetic events, identify complex genetic exchanges involving more than two organisms, and find statistical patterns in which groups of genes were more likely to be exchanged as a set. In an investigation of the H7N9 influenza virus responsible for the April 2013 bird flu outbreak in China, their method was able to recapitulate the complex reassortment process that led to the emergent virus (figure C).
 The authors suggest that persistent homology can not only identify patterns in genetic evolution in viruses, but will also be useful in understanding evolution in nonviral species. More generally, it could be used to address the fundamental challenge of developing a more precise map of evolution at the molecular level.
The authors suggest that persistent homology can not only identify patterns in genetic evolution in viruses, but will also be useful in understanding evolution in nonviral species. More generally, it could be used to address the fundamental challenge of developing a more precise map of evolution at the molecular level.
— Chris Williams
Related publication
Chan JM, Carlsson G, Rabadan R. Topology of viral evolution . Proc Natl Acad Sci USA. 2013 Nov 12;110(46): 18566-71.