News
New Algorithm Accurately Predicts Drug Mechanism of Action
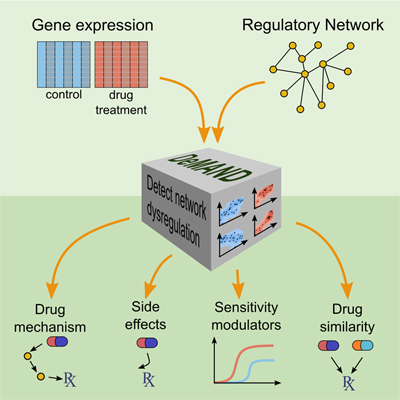
By analyzing drug-induced changes in disease-specific patterns of gene expression, a new algorithm called DeMAND identifies the genes involved in implementing a drug's effects. The method could help predict undesirable off-target interactions, suggest ways of regulating a drug's activity, and identify novel therapeutic uses for FDA-approved drugs, three critical challenges in drug development.
Researchers in the Columbia University Department of Systems Biology have developed an efficient and accurate method for determining a drug’s mechanism of action — the cellular machinery through which it produces its pharmacological effect. Considering that most drugs, including widely used ones, act in ways that are not completely understood at the molecular level, this accomplishment addresses a key challenge to drug development. The new approach also holds great potential for improving drugs’ effectiveness, identifying better combination therapies, and avoiding dangerous drug-induced side effects.
According to Andrea Califano, the Clyde and Helen Wu Professor of Chemical Systems Biology and co-senior author on the study, “This new methodology makes it possible for the first time to generate a genome-wide footprint of the proteins that are responsible for implementing or modulating the activity of a drug. The accuracy of the method has been the most surprising result, with up to 80% of the identified proteins confirmed by experimental assays.”
During a drug’s development, scientists design compounds that bind to one or more specific targets within a cell, producing a desired effect. However, when the drug enters the human body it becomes part of an incredibly complex system, and can interact with other molecules in ways that are hard to predict. Current binding assays, such as those to assess the effect on protein classes such as protein kinases, lack the ability to identify the full repertoire of proteins that may be affected by or may modulate the activity of a drug. Complicating matters further is the fact that what a drug does can vary from cell type to cell type. As a result, toxicity can occur when drugs that target drivers of disease in one cell type perform unexpected functions in others. Too often, it is these unanticipated side effects that prevent a promising drug candidate from making it into the clinic.
To address this problem, the authors used a combination of experimental and computational methods to develop a new algorithm, called Detecting Mechanism of Action by Network Dysregulation (DeMAND). DeMAND builds on over a decade of work in the Califano Lab that has produced many widely used algorithms for modeling networks of molecular interactions in specific cell types.
To predict gene products involved in a drug’s mechanism of action, the scientists first use methods developed previously in the Califano Lab to generate molecular interaction models (called interactomes) that are specific to the disease being studied. They then expose diseased cells to a drug of interest and measure gene expression changes by RNA sequencing. Measurements are taken at multiple time points and at multiple drug concentrations, creating a precise picture of the drug’s effects.
Using concepts from information theory, DeMAND then assesses how each interaction in the cell-specific interactome is affected by the drug activity, leading to a precise, deterministic picture of the drug’s mechanism of action. Specifically, for every pair of genes representing an interaction, the algorithm computes the level of dysregulation introduced between them following exposure to the drug. It then identifies proteins participating in the most significant number of dysregulated interactions, hypothesizing that these are the ones most likely to play a key role in the drug’s mechanism of action.
DeMAND identifies not just the drug's direct target but also other gene products that are involved in implementing its effect or modulating its activity.
Other methods exist for identifying how a drug binds to its direct target, although these tend to be very labor intensive and capable only of identifying targets to which a compound binds most strongly. DeMAND constitutes an important leap forward because it identifies not just the drug’s direct target but also other gene products that are involved in implementing its effect or modulating its activity. This systems biology approach, then, provides additional context for the drug’s activity that can be used to anticipate off-target effects.
Despite these advantages, DeMAND is also much more efficient than previous methods. As Dr. Califano explains, “One can now elucidate the mechanism of action of a drug in many different tissues by just generating a very small number of perturbational profiles (6 or more). This dramatically increases our ability to assess both on-target and off-target effects that might be tissue-specific.”
DeMAND’s predictions proved to be very accurate. The researchers report that when they exposed human diffuse B-cell lymphoma cells to a panel of drugs, the algorithm identified previously documented targets for 70% of the tested compounds. Moreover, it also predicted many never-before-identified proteins involved in drug mechanisms of action, more than 80% of which were either already known in the literature or were validated by follow-up experiments. These findings suggest that DeMAND is capable of providing reliable new insights into drug mechanism of action that can be tested immediately.
Furthermore, the algorithm makes it possible to compare mechanisms of action between two compounds, and to identify compounds with similar pharmacological effect. It determined, for example, that the mechanism of action for altretamine, an anti-cancer drug with no known targets or effectors, was very similar to that of sulfasalazine, another drug whose pathway activity is known. Although these two drugs were previously unrelated, DeMAND showed that altretamine could potentially be used as an alternative drug for patients who do not benefit from sulfasalazine.
Co-senior author Mukesh Bansal sees great potential in this approach, saying, “DeMAND can accelerate the drug discovery process by unraveling the underlying mechanisms of action of new compounds. Our findings about altretameine also show that it can determine novel therapeutic applications for existing FDA approved drugs. This will dramatically reduce the cost of drug development and provide a huge potential to benefit patients.”
— Chris Williams
Related publication
Woo JH, Shimoni Y, Yang WS, Subramaniam P, Iyer A, Nicoletti P, Rodríguez Martínez M, López G, Mattioli M, Realubit R, Karan C, Stockwell BR, Bansal M, Califano A. Elucidating compound mechanism of action by network perturbation analysis . Cell. 2015 Jul 16;162(2):441-51.