News
New Tool for Visualizing High-Dimensional Single-Cell Data Reveals Phenotypic Heterogeneity in Leukemia
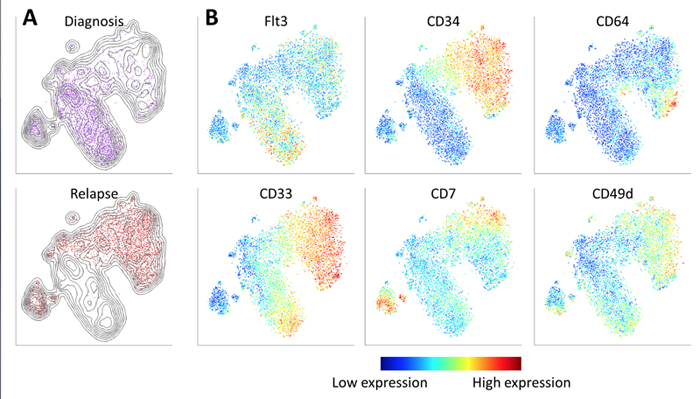
viSNE reveals the progression of cancer in a sample of cells taken from a patient with acute myeloid leukemia. Cells are colored according to intensity of expression of the indicated cell markers, enabling the comparison of expression patterns before and after relapse. For example, Fit3 is expressed primarily in the diagnosis sample, while CD34 emerges in the relapse sample.
Researchers in the Columbia Initiative in Systems Biology have developed a computational method that enables scientists to visualize and interpret high-dimensional data produced by single-cell measurement technologies such as mass cytometry. The method, called viSNE (visual interactive Stochastic Neighbor Embedding), has just been published in the online edition of Nature Biotechnology. It has particular relevance to cancer research and therapeutics. As Columbia University Medical Center reports:
Researchers now understand that cancer within an individual can harbor subpopulations of cells with different molecular characteristics. Groups of cells may behave differently from one another, including in how they respond to treatment. The ability to study single cells, as well as to identify and characterize subpopulations of cancerous cells within an individual, could lead to more precise methods of diagnosis and treatment.
“Our method not only will allow scientists to explore the heterogeneity of cancer cells and to characterize drug-resistant cancer cells, but also will allow physicians to track tumor progression, identify drug-resistant cancer cells, and detect minute quantities of cancer cells that increase the risk of relapse,” said co-senior author Dana Pe’er, associate professor of biological sciences and systems biology at Columbia.
The other co-senior author is Garry P. Nolan, professor of microbiology & immunology at Stanford.
The viSNE software can analyze measurements of dozens of molecular markers. In the maps that result, the distance between points represents the degree of similarity between single cells. The maps can reveal clearly defined groups of cells with distinct behaviors (e.g., drug resistance) even if they are only a tiny fraction of the total population. This should enable the design of ways to physically isolate and study these cell subpopulations in the laboratory.
Although the algorithm underlying the method is complex, Dr. Pe’er expects that all researchers, no matter their level of mathematical expertise, will be able to use viSNE.
To demonstrate the software’s utility, Dr. Pe’er and her colleagues used mass cytometry and viSNE to study bone marrow cells from patients with acute myeloid leukemia. Currently, clinicians can incorporate at most 4 to 8 markers to assess the cells. Because mass cytometry and viSNE can incorporate many more markers, viSNE is able to identify more subtle differences between cells. Using the algorithm, Dr. Pe’er and her colleagues were able to reveal previously unrecognized heterogeneity in the bone marrow cells they studied.
The researchers also showed that viSNE could detect minimal residual disease (MRD) — extremely small quantities of cancer cells that persist after chemotherapy and raise the risk of recurrence. “In blinded tests, we were able to find as few as 20 cancer cells out of tens of thousands of healthy cells,” said Dr. Pe’er. Such a small quantity of cells is extremely difficult to detect, even by the most experienced pathologist.
“The ability to detect MRD is critical for curing cancer,” added Dr. Pe’er. “Eliminating even 99.9 percent of a tumor doesn’t bring about a cure. You have to be able to find, and then eliminate, the tiny populations of cells that can survive therapy and lead to disease relapse.”