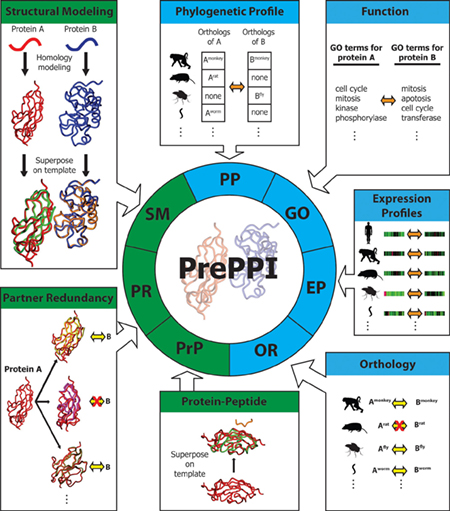
PrePPI predicts the likelihood that two proteins A and B are capable of interacting based on their similarities to other proteins that are known to interact. This requires integrating structural data (green) as well as other kinds of information (blue), such as evidence of protein co-activity in other species as well as involvement in similar cellular functions. PrePPI now offers a searchable database of unprecedented scope, constituting a virtual interactome of all proteins in human cells. (Image courtesy of eLife.)
The molecular machinery within every living cell includes enormous numbers of components functioning at many different levels. Features like genome sequence, gene expression, proteomic profiles, and chromatin state are all critical in this complex system, but studying a single level is often not enough to explain why cells behave the way they do. For this reason, systems biology strives to integrate different types of data, developing holistic models that more comprehensively describe networks of interactions that give rise to biological traits.
Although the concept of an interaction network can seem abstract, at its foundation each interaction is a physical event that takes place when two proteins encounter one another, bind, and cause a change that affects a cell’s activity. In order for this to take place, however, they need to have compatible shapes and physical properties. Being able to predict the entire universe of possible pairwise protein-protein interactions could therefore be immensely valuable to systems biology, as it could both offer a framework for interpreting the feasibility of interactions proposed by other methods and potentially reveal unique features of networks that other approaches might miss.
In a 2012 paper in Nature, scientists in the laboratory of Barry Honig first presented a landmark algorithm and database they call PrePPI (Predicting Protein-Protein Interactions). At the time, PrePPI used a novel computational strategy that deploys concepts from structural biology to predict approximately 300,000 protein-protein interactions, a dramatic increase in the number of available interactions when compared with experimentally generated resources.
Since then, the Honig Lab has been working hard to improve PrePPI’s scope and usefulness. In a paper recently published in eLife they now report on some impressive developments. With enhancements to their algorithm and the incorporation several new types of data into its analysis, the PrePPI database now contains more than 1.35 million predictions of protein-protein interactions, covering about 85% of the entire human proteome. This makes it the largest resource of its kind. In parallel with these improvements, the investigators have also begun to apply PrePPI in new ways, using the information it contains to provide new kinds of insights into the organization and function of protein interaction networks.