News
Database of Protein-Protein Interactions Opens New Possibilities for Systems Biology
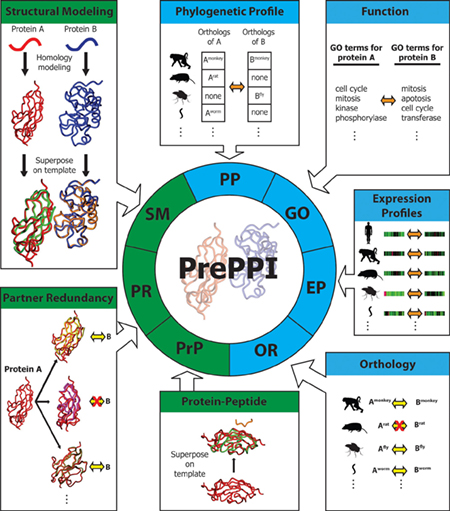
PrePPI predicts the likelihood that two proteins A and B are capable of interacting based on their similarities to other proteins that are known to interact. This requires integrating structural data (green) as well as other kinds of information (blue), such as evidence of protein co-activity in other species as well as involvement in similar cellular functions. PrePPI now offers a searchable database of unprecedented scope, constituting a virtual interactome of all proteins in human cells. (Image courtesy of eLife.)
The molecular machinery within every living cell includes enormous numbers of components functioning at many different levels. Features like genome sequence, gene expression, proteomic profiles, and chromatin state are all critical in this complex system, but studying a single level is often not enough to explain why cells behave the way they do. For this reason, systems biology strives to integrate different types of data, developing holistic models that more comprehensively describe networks of interactions that give rise to biological traits.
Although the concept of an interaction network can seem abstract, at its foundation each interaction is a physical event that takes place when two proteins encounter one another, bind, and cause a change that affects a cell’s activity. In order for this to take place, however, they need to have compatible shapes and physical properties. Being able to predict the entire universe of possible pairwise protein-protein interactions could therefore be immensely valuable to systems biology, as it could both offer a framework for interpreting the feasibility of interactions proposed by other methods and potentially reveal unique features of networks that other approaches might miss.
In a 2012 paper in Nature, scientists in the laboratory of Barry Honig first presented a landmark algorithm and database they call PrePPI (Predicting Protein-Protein Interactions). At the time, PrePPI used a novel computational strategy that deploys concepts from structural biology to predict approximately 300,000 protein-protein interactions, a dramatic increase in the number of available interactions when compared with experimentally generated resources.
Since then, the Honig Lab has been working hard to improve PrePPI’s scope and usefulness. In a paper recently published in eLife they now report on some impressive developments. With enhancements to their algorithm and the incorporation several new types of data into its analysis, the PrePPI database now contains more than 1.35 million predictions of protein-protein interactions, covering about 85% of the entire human proteome. This makes it the largest resource of its kind. In parallel with these improvements, the investigators have also begun to apply PrePPI in new ways, using the information it contains to provide new kinds of insights into the organization and function of protein interaction networks.
“Ultimately what we are doing,” Honig says, “is to use protein structures in ways they’re not normally used. We are leveraging the large amount of structural data that now exists and are finding that it gives us the opportunity to answer questions that couldn’t otherwise be addressed.” As the new paper reports, this includes providing new methods for interpreting genetic data, annotating protein function, and identifying promising new targets for treating human disease.
The new and improved PrePPI
PrePPI is not the first approach for predicting protein-protein interactions, although its use of structural modeling, combined with several other kinds of information, distinguishes it from others. The algorithm’s design relies on the observation, documented in the Protein Data Bank and elsewhere, that many proteins contain structural similarities. It also takes inspiration from the widely used bioinformatics tool BLAST — which identifies similarities in protein sequence to identify proteins with similar functions — constituting what the Honig Lab refers to as a “structural BLAST.” Here, they use approaches called structural alignment and homology modeling to identify structural similarities in different proteins that would suggest that they have similar binding properties.
In practice this means that if researchers want to know whether two proteins A and B interact, they first develop structural models of the proteins. They then look for what they call “structural neighbors,” A′ and B′, that have known structures and interaction profiles that are similar to those of A and B. If A′ and B′ are known to interact, this provides an important clue that A and B might also interact at the same interface. (To learn more about the theory at the core of the original PrePPI, see Uniting Structural and Systems Biology: An Interview with Barry Honig.)
Once this structural compatibility of protein pairs was scored, the original PrePPI also incorporated additional information to develop more confidence that the predicted interaction was actually real. This included considering the phylogenetic profile of proteins A and B (that is, whether they are typically found together in the same animal species), and their similarities in Gene Ontology (GO) analysis, a database that describes the functions of specific proteins in the cell. The newest generation of PrePPI enhances the utility of these data points by incorporating several new features. This includes scoring orthology (the likelihood that A and B interact if the template proteins A′ and B′ are known to interact in other species), expression profile (similarity between the gene expression patterns in proteins A and B and those of their orthologs in other model organisms), partner redundancy (how many close structural neighbors of protein A there are that protein B is known to interact with), and protein-peptide relationships (whether there are known interactions between a structured domain on A and an unstructured peptide in B that contains a short sequence motif that is known to interact with A).
At the end of the pipeline, these various scores are integrated into an equation that calculates a likelihood ratio that proteins A and B interact. Using the Department of Systems Biology’s high-performance computing cluster, the Honig Lab tested potential pairwise protein-protein actions across the entire human genome. The resulting PrePPI database now contains approximately 1.35 million high-confidence predictions. This resource is available publicly through the Honig Lab website.
The new paper reports that in addition to quadrupling the number of predicted interactions contained in the first release of PrePPI, the upgraded version’s integration of new kinds of information also improves prediction performance, to the degree that its capabilities now rival those of high-throughput experimentation. The researchers anticipate that this will continue to improve as they incorporate new data and update the database annually.
In effect, the increased scope of PrePPI means that it now provides a virtual genome-wide interactome of protein-protein interactions. By querying the database, it becomes possible to determine a probability that almost any potential interaction of interest actually occurs in the cell.
“We feel we have a very unique resource,” Honig says, “and now we would like it to be more accessible and more usable for others. We’re also beginning to apply PrePPI in various ways, because the more we use it and the more successes we have with it, the more others in the research community will want to use it themselves.”
Putting PrePPI to work against K-Ras and other cancer proteins
One area in which the Honig Lab is interested in applying PrePPI is to identify new strategies for inhibiting the destructive effects of the protein K-Ras. Mutated K-Ras is known to play a role in a variety of cancers, but researchers have struggled to find effective therapeutics because its properties render it virtually undruggable. Taking a different approach, Honig and his colleagues are interested in determining whether interfering with the proteins that interact with K-Ras could offer other opportunities for disrupting its activity.
In a recent collaboration with Timothy Wang in Columbia University Medical Center’s Department of Digestive and Liver Diseases, for example, the Honig Lab used PrePPI to investigate whether a protein kinase called Dclk1 is likely to interact with K-Ras. Based on other research in his lab, Wang postulated that Dclk1 might activate K-Ras in pancreatic cancer, and when they looked in PrePPI, they found that the Dclk1/K-Ras interaction indeed has a high score. In addition, the query revealed a strong structural similarity between a specific domain in Dclk1 and that of another protein called RalGDS, whose interaction with K-Ras has been confirmed experimentally using protein crystallography. Based on this and other findings, the Wang Lab showed that Dclk1 is a marker of pancreatic cancer progenitor cells. Because the protein is a targetable kinase for which structural information is now known, the finding could also offer a new strategy for preventing those progenitors from turning malignant.
As part of a new grant from the National Institute of General Medical Sciences (NIGMS), the Honig Lab intends to pursue its interest in K-Ras and other proteins that are important in cancer. By using PrePPI to develop interactomes within which these proteins function, their goal is to provide a comprehensive map of the protein-protein interactions involved in cancer signaling pathways. Systematically building such interactomes for all known cancer proteins, they anticipate, could become a valuable resource for cancer researchers everywhere.
The Honig Lab is also working closely with the laboratory of Andrea Califano, who recently published an algorithm called VIPER, which infers protein activity by looking at changes in expression of the genes those proteins regulate. By integrating PrePPI, VIPER, and other tools for network analysis in the Califano Lab, the researchers anticipate being able to gain a more precise understanding of the key proteins responsible for driving cancer phenotypes, and to develop robust models of the interaction networks surrounding them.
Classifying SNPs using structural information
One other approach that the Honig Lab is exploring is to integrate protein-protein interaction data with genetic data. Using next-generation sequencing and statistical analysis, geneticists can identify subtle differences in nucleotide sequences that distinguish healthy individuals from those with disease. However, because alterations called single nucleotide polymorphisms (SNPs) are often rare and difficult to distinguish from normal genetic diversity, the field struggles to tell whether they actually cause disease or are merely associated with it.
The eLife paper proposes that structural biology could help address this challenge. The Honig Lab compiled a complete list of SNPs included in two publicly available databases and asked PrePPI which alterations were likely to be located at protein-protein interfaces. Their analysis showed that the likelihood of a disease-associated SNP to be located at a protein-protein interface is much higher than one would expect by chance. In addition, benign SNPs included in the 1000 Genomes Project database were significantly underrepresented at protein interfaces.
The paper argues that these findings support a growing body of evidence that disruptions of protein-protein interactions play important roles in causing disease. Moreover, it indicates that the information contained in PrePPI could be useful in helping to distinguish disease-causing SNPs from those that have no effect.
Using protein-protein interactions to annotate protein function
In addition to identifying disease-associated mutations, PrePPI can also be used to gain a better understanding of the cellular processes in which a gene or protein is involved. Similarly to how the Honig Lab took the BLAST concept and applied it to protein structure, they have also begun using PrePPI within a gene set enrichment analysis (GSEA) framework.
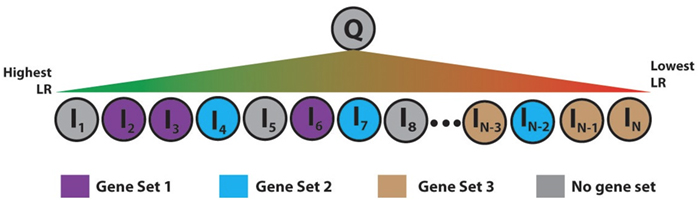
Using PrePPI for gene set enrichment analysis. To infer the function of a particular protein, Q, the Honig Lab places all proteins in the human proteome, li, in a list and sorts them according to the interaction likelihood ratio between li and Q. They then search for gene sets associated with a given Gene Ontology annotation that is enriched among the high-scoring interactors of Q. In the example, Gene Set 1 would be enriched whereas Gene Sets 2 and 3 would not be, since the proteins in those sets are either evenly distributed throughout the ranked list or clustered with proteins that are unlikely to interact with Q. The paper reports on top ranked gene sets found for BRCA1 and PEX2. (Image courtesy of eLife.)
For each protein, they queried PrePPI to construct a list of the proteins whose scores make them most likely to interact with it, and then, using GSEA, looked for the GO terms associated with each. They found that the top-ranked gene sets that PrePPI predicted accurately reflected their function, as documented in a resource called the Molecular Signatures Database (mSigDB). Moreover, through the automatic computational method made possible by PrePPI, they predicted the functions of approximately 2,000 additional proteins whose functions were previously unknown.
Honig cautions that the interactions and functions predicted using PrePPI should not necessarily be assumed as fact. Nevertheless, his lab’s tests so far indicate that they are largely reliable. “PrePPI is based on statistical analysis and not experiment, which is really the gold standard,” he explains. “What we’re ultimately trying to do with these methods is to generate hypotheses that can be cross-referenced with other computational and experimental methods. We’re excited because the number of interactions that PrePPI finds is unprecedented in scope, and so our hope is that it will help systems biologists and other biomedical researchers who do not typically look at structure to be able to incorporate information about this essential layer of activity into their investigations."
— Chris Williams
Related publications
Garzón JI, Deng L, Murray D, Shapira S, Petrey D, Honig B. A computational interactome and functional annotation for the human proteome. Elife. 2016 Oct 22;5. pii: e18715.
Westphalen CB, Takemoto Y, Tanaka T, Macchini M, Jiang Z, Renz BW, Chen X, Ormanns S, Nagar K, Tailor Y, May R, Cho Y, Asfaha S, Worthley DL, Hayakawa Y, Urbanska AM, Quante M, Reichert M, Broyde J, Subramaniam PS, Remotti H, Su GH, Rustgi AK, Friedman RA, Honig B, Califano A, Houchen CW, Olive KP, Wang TC. Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell. 2016 Apr 7;18(4):441-55.
Chen TS, Petrey D, Garzon JI, Honig B. Predicting peptide-mediated interactions on a genome-wide scale. PLoS Comput Biol. 2015 May 4;11(5):e1004248.
Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012 Oct 25;490(7421):556-60.
Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, Califano A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016 Aug;48(8):838-47.