News
Graduate Students Invent Technique for Reprogramming Translation
 MD/PhD students Andrew Anzalone and Sakellarios Zairis combined approaches based in chemical biology, synthetic biology, and computational biology to develop a new method for protein engineering.
MD/PhD students Andrew Anzalone and Sakellarios Zairis combined approaches based in chemical biology, synthetic biology, and computational biology to develop a new method for protein engineering.
The ribosome is a reliable machine in the cell, precisely translating the nucleotide code carried by messenger RNAs (mRNAs) into the polypeptide chains that form proteins. But although the ribosome typically reads this code with uncanny accuracy, translation has some unusual quirks. One is a phenomenon called -1 programmed ribosomal frameshifting (-1 PRF), in which the ribosome begins reading an mRNA one nucleotide before it should. This hiccup bumps translation “out of frame,” creating a different sequence of three-nucleotide-long codons. In essence, -1 PRF thus gives a single gene the unexpected ability to code for two completely different proteins.
Recently Andrew Anzalone, an MD/PhD student in the laboratory of Virginia Cornish, set out to explore whether he could take advantage of -1 PRF to engineer cells capable of producing alternate proteins. Together with Sakellarios Zairis, another MD/PhD student in the Columbia University Department of Systems Biology, the two developed a pipeline for identifying RNA motifs capable of producing this effect, as well as a method for rationally designing -1 PRF “switches.” These switches, made up of carefully tuned strands of RNA bound to ligand-sensing aptamers, can react to the presence of a specific small molecule and reliably modulate the ratio in the production of two distinct proteins from a single mRNA. The technology, they anticipate, could offer a variety of exciting new applications for synthetic biology. A paper describing their approach and findings has been published in Nature Methods.
A new twist on an old technology
Although scientists had been aware of -1 PRF for some time, it was unclear when Anzalone began his project what specific features in RNA could cause ribosome slippage. His first step, then, was to develop a framework for discovering them.
His technique is based on mRNA display, invented 15 years ago by Nobel laureate Jack Szostak to isolate proteins with desired functions. In this method, libraries of up to 100 trillion unique oligonucleotides (short strings of DNA) are synthesized with randomized nucleotides scattered throughout the sequence. Each DNA library member is then transcribed to RNA and tagged with an antibiotic called puromycin, which during in vitro protein synthesis covalently binds the RNA to the resulting protein. Using high-throughput sequencing, it then becomes possible to retroactively identify the RNA sequences that produced proteins of interest.
In order to better understand how frameshifting occurs during translation, Anzalone subjected his initial pool of oligonucleotides (268 million unique sequences) to a series of in vitro selection experiments. In each case he kept sequences that induced frameshifting and eliminated those that did not. mRNA display could discriminate between the two groups, Anzalone realized, because in order for puromycin to attach an RNA to its resulting polypeptide, the ribosome must translate all the way to the end of the RNA. He reasoned that if a frameshift did not occur the ribosome would hit a stop codon in an mRNA. If a frameshift took place, however, the stop signal would not longer exist, making it possible for the ribosome to translate the complete RNA sequence. As a result, he could use the bound proteins as tags to enrich RNA sequences that cause a frameshift.
“I found myself in a unique position,” Anzalone explains, “where I was using mRNA display in a way no one else had thought of because it wasn’t what it was designed for. Luckily, it happened to be very useful for what I was doing.” After three rounds of selection, his pool of RNAs was highly enriched for those associated with -1 PRF.
Finding frameshift-inducing structures
The next step was to use high-throughput sequencing to see how the RNA molecules most associated with the frameshift differed from the larger pool of sequences present before selection. Because Anzalone was trained as a chemist and not a computational biologist, however, he needed a mathematical framework for interpreting the unique dataset he had created.
Around this time he was discussing his project with Sakellarios Zairis, a friend and classmate in Columbia’s MD/PhD program, and the two realized that collaborating could lead to a solution. As a member of Raul Rabadan’s lab, Zairis had been working on methods for quantifying patterns of evolution in tumors from longitudinal sequencing data. Although -1 PRF exists in a completely different biological domain from cancer genetics, he recognized certain similarities in the problem. “I was already thinking about how to construct useful parameter spaces for analyzing genome evolution under strong selection,” he recalls. “When Andrew first started talking about his dataset, I wondered how we could represent it in a way that would allow you to explore it intuitively.”
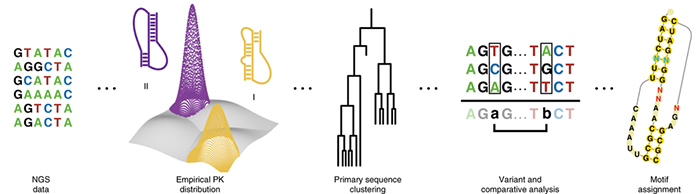
Following selection for -1 PRF, Zairis grouped them into pseudoknot (PK) families, analyzed for sequences enriched following selection, and then clustered them based on primary sequence identity. This makes it possible to compare motifs, or identify changes in nucleotide sequence that lead to stable structures.
Hypothesizing that structural features in RNA must be responsible for bumping translation out of frame, Zairis began learning about three-dimensional RNA features called pseudoknots and hairpins, in which linear sequences of nucleotides curl into loop-like shapes due to interactions among the nucleotides. The key insight came when he realized that the space of pseudoknots accessible to the RNAs in the selection library had seven key structural components, each of which can be summarized by an integer. Moreover, because these structures require that nucleotides on the same RNA bind to one another to keep the molecule stable, each of these parameters has a limited range of possibilities, making the space of secondary structures far more constrained and tractable than the space of primary sequences. To analyze the dataset, then, Zairis developed a mathematical pipeline capable of interpreting RNA sequencing data to identify families of pseudoknot structures that were most feasible. (Visit Github to see Zairis’s frameshift analyzis pipeline.)
Using the Department of Systems Biology’s high-performance computing cluster, Zairis compared the distributions of pseudoknot geometries seen before Anzalone’s selection experiments to those seen after the selection for -1 PRF. In the end, instead of needing to contend with 268 million possible pseudoknots based on sequence alone, his reduced representation condensed the dataset to approximately 2000 possible structural families, making it possible to calculate the top motifs and interpret their biological function. Once they identified the geometries that induced frameshifting, they went a step further, identifying the particular nucleotide preferences within those geometries, revealing instances of single substitutions that have strong effects on pseudoknot fitness.
In caffeine-fueled all night working sessions, the friends became excited by what the algorithm was unearthing. Zairis remembers the experience fondly, saying, “I would ask, ‘What do you think of this geometry?’ and Andrew’s eyes would light up and he’d say, ‘I recognize that geometry. That’s an important pseudoknot I’ve seen in the literature.’” In this way, they quickly gained a clearer picture of the specific RNA motifs most capable of producing -1 PRF. Once they understood the basic biology, Anzalone could then begin incorporating these findings into molecular tools capable of reliably producing specific frameshifts.
Engineering a -1 PRF inducing riboswitch
Because of his interests in chemical biology, Anzalone’s goal all along had been to design biochemical structures capable of changing protein translation in the presence of specific small molecules. Such structures, called riboswitches, had been designed in the past, although none had been used to induce frameshifting of the sort he had been investigating.
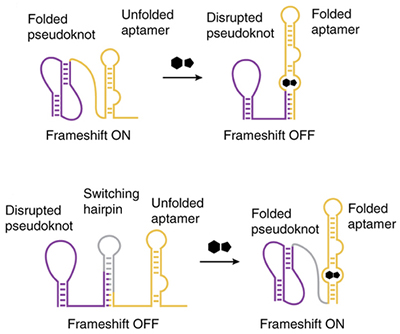
Two kinds of -1 PRF switches: In the first case, a pseudoknot that stimulates -1 PRF is energetically dominant, producing high frameshift levels. When a specific small molecule is present, the aptamer folds, disrupting the pseudoknot structure. In the second, a switching hairpin disrupts the pseudoknot and frameshifting. In the presence of ligand, the aptamer folds and destabilizes the switching hairpin, allowing the pseudoknot to induce a frameshift.
Using a rational design approach incorporating the results of the aforementioned computational studies, Anzalone undertook a series of experiments in which he bound -1 PRF-inducing pseudoknots to small molecule-sensing aptamers. When a specific target small molecule is present, the aptamer binds to it and changes shape. This also changes the geometry of the pseudoknot, subsequently turning -1 PRF on or off. As he optimized the system over a series of experiments, Anzalone was pleased to discover that the ratio of frameshifted proteins to non-frameshifted proteins resulting from reactions at the ribosome was very pronounced and consistent in response to the addition of a small molecule.
By combining various riboswitches, Anzalone also showed that this approach could enable a kind of biological computation, creating more complex logic gates representing AND or OR functions that could be contained on a single mRNA transcript. As a proof of principle, he designed a riboswitch for regulating apoptosis in yeast that could consistently change the ratio of viable cells to cells that undergo programmed cell death based on application of the drugs theophylline and neomycin.
Anzalone argues that riboswitches that leverage -1 PRF could offer some unique opportunities for synthetic biology. “A lot of things in biology are not about turning something on or off, but about maintaining a balance between two different regulators with opposing functions,” Anzalone explains. “I realized that by using our method to set the stoichiometry and even switch between stoichiometries based on the presence of a small molecule, you could basically switch the phenotype of a cell.”
Other potential applications, he suggests, could include perturbing biological systems to understand underlying networks, incorporating labeling technologies to track individual cells, or using frameshifting motifs as sensors of specific activities inside the cell. Both the experimental and computational approaches that Anzalone and Zairis created are designed to adaptable for a wide range of biological settings.
Anzalone's advisor, Virginia Cornish, expresses excitement at the achievement, saying, "For a long time my laboratory has been interested in engineering translation to work with unnatural amino acid building blocks, as well as in synthetic biology. So it was wonderful when Andrew came to me with the idea of using ribosomal frameshifting to bring these two areas together. As an advisor there is nothing more satisfying that to see a graduate student achieve that level of intellectual independence."
“When I saw the results,” says Raul Rabadan, Zairis's advisor, “I was very impressed. It’s a great example of the kinds of collaboration that can happen in the Department of Systems Biology, where clever and driven students from two labs with very different interests can come together to do something quite unusual.”
— Chris Williams
Related publication
Anzalone AV, Lin AJ, Zairis S, Rabadan R, Cornish VW. Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches. Nat Methods. 2016 May;13(5):453-458.