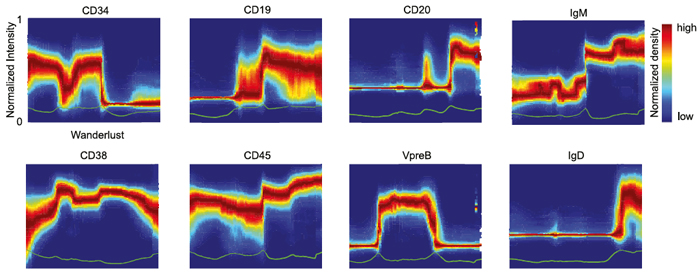
A new algorithm called Wanderlust uses single-cell measurements to detect how marker expression changes across development.
In a new paper published in the journal Cell, a team of researchers led by Dana Pe’er at Columbia University and Garry Nolan at Stanford University describes a powerful new method for mapping cellular development at the single cell level. By combining emerging technologies for studying single cells with a new, advanced computational algorithm, they have designed a novel approach for mapping development and created the most comprehensive map ever made of human B cell development. Their approach will greatly improve researchers’ ability to investigate development in cells of all types, make it possible to identify rare aberrations in development that lead to disease, and ultimately help to guide the next generation of research in regenerative medicine.
Pointing out why being able to generate these maps is an important advance, Dr. Pe’er, an associate professor in the Columbia University Department of Systems Biology and Department of Biological Sciences, explains, “There are so many diseases that result from malfunctions in the molecular programs that control the development of our cell repertoire and so many rare, yet important, regulatory cell types that we have yet to discover. We can only truly understand what goes wrong in these diseases if we have a complete map of the progression in normal development. Such maps will also act as a compass for regenerative medicine, because it’s very difficult to grow something if you don’t know how it develops in nature. For the first time, our method makes it possible to build a high-resolution map, at the single cell level, that can guide these kinds of research.”