News
Mechanism of Kidney Transplant Tolerance Discovered
After identifying T cell clones that react against donated kidney tissue in vitro, new computational methods developed in Yufeng Shen's Lab are used to track their frequency following organ transplant. The findings can help to predict transplant rejection or tolerance.
When a patient receives a kidney transplant, a battle often ensues. In many cases, the recipient’s immune system identifies the transplanted kidney as a foreign invader and mounts an aggressive T cell response to eliminate it, leading to a variety of destructive side effects. To minimize complications, many transplant recipients receive drugs that suppress the immune response. These have their own consequences, however, as they can lead to increased risk of infections. For these reasons, scientists have been working to gain a better understanding of the biological mechanisms that determine transplant tolerance and rejection. This knowledge could potentially improve physicians’ ability to predict the viability of an organ transplant and to provide the best approach to immunosuppression therapy based on individual patients’ immune system profiles.
Yufeng Shen, an assistant professor in the Columbia University Department of Systems Biology and JP Sulzberger Columbia Genome Center, together with Megan Sykes, director of the Columbia Center for Translational Immunology at the Columbia University College of Physicians and Surgeons, recently took an encouraging step toward this goal. In a paper published in Science Translational Medicine, they report that the deletion of specific donor-resistant T cell clones in the transplant recipient can support tolerance of a new kidney. Critical to this discovery was the development of a new computational genomics approach by the Shen Lab, which makes it possible to track how frequently rare T cell clones develop and how their frequencies change following transplantation. The paper suggests both a general strategy for understanding the causes of transplant rejection and a means of identifying biomarkers for predicting how well a transplant recipient will tolerate a new kidney.
Tracking the evolution of a T cell army
Because of their specialized function within the immune system, T cells have unique genetic properties. One defining feature is the T-cell receptor. This protein is located on the surface of the T cell and recognizes “foreign” agents from outside the body. As T cells mature, a somatic process called V(D)J recombination produces diverse populations of clones. In aggregate, these clones present a broad spectrum of T cell receptors, enabling the immune system to respond to many different pathogens. “It’s like having a standing army,” Shen explains. “You want a wide variety of T cell receptors even before a pathogen enters the body so that you have soldiers ready to respond to it.”
This constant state of preparedness becomes a problem, however, following an organ transplant, as a subset of T cell clones reacts much as it would to infection. To understand the mechanism of transplant rejection, the investigators wanted to determine which clones are responsible. To do so, they utilized an ex vivo assay called mixed lymphocyte reaction (MLR). In this experiment, blood from the recipient and a small amount of donor tissue are placed together in a tube before the transplant. When brought together in this way, lymphocytes — a class of immune cells that includes T cells — react to the tissue in the same way one would expect they would in the body. The investigators then perform bulk deep sequencing on the sample to identify T cell clones that react against the donor tissue. This is possible because the genetically diverse coding regions for the T cell receptor serve as a kind of “barcode” for each clone sequence.
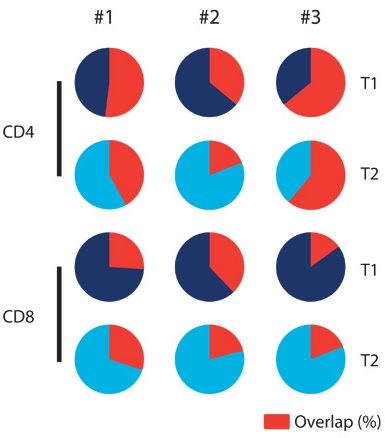
Pie charts showing the relative overlap of the summed frequencies of donor-reactive clones over time in three healthy controls. Each circle represents the cumulative frequency of all donor-reactive clones identified in the sample. The red segment shows the percentage of that total frequency arising from donor-reactive clones identified at both T1 and T2.
In the research described in the paper, kidney transplants were then performed on six patients. Two utilized conventional transplant methods, while four used an experimental technique that Dr. Sykes developed called combined kidney and bone marrow transplantation (CKBMT). In this procedure, the kidney transplant is accompanied by the transplantation of bone marrow from the donor to the recipient. Once incorporated into the recipient’s body, the donated bone marrow begins generating its own T cells, leading to the development of a hybrid immune system that can recognize the donated organ as “self.” Following kidney transplant in all six patients, blood was drawn at 6, 12, and 18 months, and additional deep sequencing analyses were performed at each time point. This enabled the investigators to determine how the donor-reactive T cell clones changed due to interactions between the recipient’s immune system and the donated kidney.
The behavior of the donor-reactive T cell clones in these follow-up sequencing experiments provides clues about how well the recipient’s immune system tolerates the foreign kidney. If there is an expansion in the frequency of donor-reactive clones following transplantation, the researchers hypothesized that there should be a reaction against the donated organ, usually indicating transplant rejection. In the study, this phenomenon occurred in the two patients who received a conventional kidney transplant without bone marrow transplant. Intriguingly, the investigators found that in 3 of the 4 patients receiving CKBMT, the frequency of donor-reactive clones identified in the original MLR experiment decreased, and each patient tolerated the transplant. In the fourth patient, the number of donor-reactive cells did not decrease, and the patient rejected the kidney
These observations suggest that the new hybrid immune system can initiate what the researchers characterize as a “deletion mechanism” that actively destroys or suppresses the expansion of donor-reactive T cells. Although this surprising finding is still under investigation, it suggests a mechanism behind transplant tolerance, and provides exciting evidence that CKBMT could offer a strategy for improving the success of kidney transplantation. Currently, the investigators are planning a clinical trial of this method.
Mapping the ocean of rare T cell clones
Essential to this promising discovery was a new statistical method the Shen Lab developed for analyzing T cell genome sequencing data. “Although the audience for the paper is mostly immunologists and transplant experts,” he remarks, “more than two-thirds of the time spent on this project focused on analyzing data. We had to develop an entirely new approach for analyzing T cell diversity from scratch.”
Because there are limits to the amount of blood that can be drawn from a patient, it is extremely difficult using standard genome analysis methods to distinguish rare T cell clones. This is because clones that appear at low frequencies become indistinguishable from statistical noise caused by inherent and unavoidable limits in the accuracy of genome sequencing technologies. Although having a larger sample of blood cells could theoretically improve statistical power, it is only clinically feasible to sample a relatively small portion of the billions of T cells and hundreds of millions of T cell clones that circulate in the body. “It’s like trying to infer properties of the ocean by taking just a drop of water,” Shen says.
Borrowing ideas from statistical physics and information theory, however, he and graduate student Boris Grinshpun devised a set of methods to quantify diversity and divergence of T cell samples. Specifically, their analysis made use of statistical methods including the Simpson diversity index and generalized entropy (GE) index, techniques for measuring the concentrations of individuals when classified into types. In other applications the GE index has been used to measure income inequality. Just as an economy is made of individuals who can be classified based on their different income levels, the T cell clones in a transplant recipient’s blood are distinguished by genomic variability in the T cell receptor region. By tracking how frequently these clonal states occur and how their frequencies change over time, the investigators can observe how the immune system is responding to the transplant. If there is a very active reaction to a donor’s tissue, the diversity of the T cell population will decrease, with high numbers of clones appearing at high frequency. If the recipient tolerates the transplant, the diversity will be maintained at a normal level, with limited amounts of expansion in the number of clones.
By using bulk sequencing to look at the ensemble properties of the T-cell clone distribution in this way and comparing them across multiple time points, Shen’s method provides a general method for tracking the dynamic properties of the recipient–transplant interaction. Identifying clonal expansion in this way could potentially help predict transplant rejection before it occurs, giving physicians evidence to support the administration of more aggressive immunotherapy. And in cases in which the analysis points to transplant tolerance, this approach could indicate when treatment with immunosuppressive drugs could be safely scaled back.
The approach used in this paper is indicative of the Shen Lab’s general interest in developing new ways to accurately identify rare clonal variants. In ongoing projects, his lab is working with the Columbia Genome Center to optimize new experimental approaches to sequence T cell receptor genes and develop related computational methods for extracting signal from noisy data sets. These approaches could have applications not just in organ transplantation, but also in cancer immunotherapy and studies of autoimmune diseases. “It’s been fascinating in this paper to be able to observe such a broad range of changes,” Shen comments, “with billions of possible genomic combinations in the T cells. In the future we think these kinds of approaches could help to improve our understanding of other diseases as well.”
— Chris Williams
Related publication
Morris H, DeWolf S, Robins H, Sprangers B, LoCascio SA, Shonts BA, Kawai T, Wong W, Yang S, Zuber J, Shen Y, Sykes M. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med. 2015 Jan 28;7(272):272ra10.