News
Promising New Targets for Treating HER2-Positive Breast Cancer
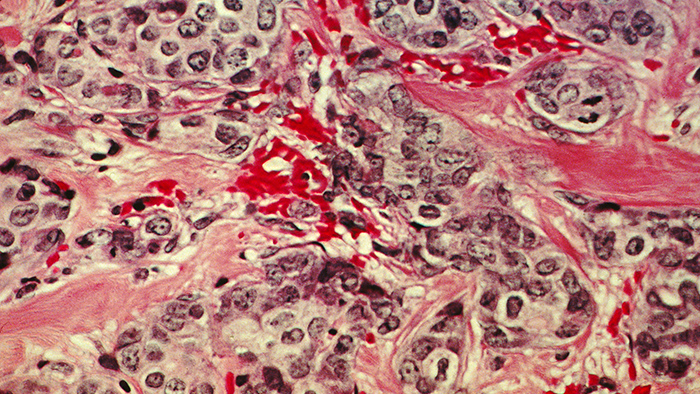
A histological slide of cancerous breast tissue. The pink "riverways" are normal connective tissue while areas stained blue are cancer cells. (Source: National Cancer Institute)
Investigators at Columbia University Medical Center and the Icahn School of Medicine at Mount Sinai have discovered a molecular signaling mechanism that drives a specific type of highly aggressive breast cancer. As reported in a paper in Genes & Development, a team led by Jose Silva and Andrea Califano determined that the gene STAT3 is a master regulator of breast tumors lacking hormone receptors but testing positive for human epidermal growth receptor 2 (HR-/HER2+). The researchers also characterized a pathway including IL-6, JAK2, STAT3, and S100A8/9 — genes already known to play important roles within the immune response — as being essential for the survival of HR-/HER2+ cancer cells. Additional tests showed that disrupting this pathway severely limits the ability of these cells to survive.
These findings are particularly exciting because the pathway the researchers identified contains multiple targets for which known FDA-approved drugs exist. The paper reports that when these drugs were tested in disease models, the cancer cells showed a dramatic response, suggesting promising strategies for the treatment of the HR-/HER2+ cancer subtype. A clinical trial is now underway to investigate the effects of these approaches in humans.
Defining a new breast cancer subtype
When a woman receives a diagnosis of breast cancer, the tumor is typically tested for the presence of human epidermal growth receptor 2 (HER2), a protein known to promote aggressive tumor growth. When a mutation in the HER2 gene is present, drugs such as trastuzumab (Herceptin) can often successfully control the tumor, although HER2-blockers are not effective against all HER2-positive tumors. Recent findings suggest that HER2-positive tumors should be sub-classified based on their hormone receptor (HR) status; that is, whether there are receptors on the surface of the cells that stimulate tumor growth in response to the presence of estrogen or progesterone. It is increasingly clear that HR-/HER2+ tumors behave differently than HR+/HER2+ tumors, including reducing the expected survival time of women with aggressive breast cancer.
An interdisciplinary approach combining pooled RNAi screens and computational algorithms identifies STAT3 as a master regulator.
In an effort to identify the molecular factors that drive the HR-/HER2+ subtype, the authors utilized a combination of innovative experimental and computational techniques. First, the Silva Lab conducted a set of genome-wide pooled RNAi screens in a model of HR-/HER2+ breast cancer. Pooled RNAi screens use short fragments of RNA to rapidly knock out the function of large numbers of individual genes, one at a time. By performing screens on both normal and HER2-transformed mammary epithelial cells, and then comparing their effects, the investigators could begin to identify genes required for the maintenance of the diseased state. To improve the reliability of their results, the Silva Lab performed three complementary RNAi screens — on tissue culture plates, in three-dimensional organ-like structures, and in immunocompromised mice. The experiments identified 355 genes that, when knocked out in all three screens, appeared to undermine the viability of HR-/HER2+ cells.
One unavoidable limitation of RNAi screens, however, is that the data they generate can be noisy, and so it was unlikely that all of these genes were truly essential for the HR-/HER2+ subtype. To address this problem, the investigators turned to computational methods as a complementary approach to narrow their focus to the most promising predictions.
Analyzing gene expression data of 359 breast tumors in The Cancer Genome Atlas (TCGA), the Califano Lab used a well-proven algorithm it developed called ARACNe to generate a model of all molecular interactions in HR-/HER2+ cells. They then applied a second algorithm called MARINa to identify genes within these interaction networks that function as master regulators. (Master regulators are transcription factors and signaling proteins that constitute bottlenecks within regulatory networks, causing a change in downstream gene expression patterns in diseased cells, when compared to normal cells of the same type.) The researchers hypothesized that targeting master regulators instead of mutated genes would offer an opportunity to stop the cancer.
When they used these regulatory models to analyze gene expression data from their model of normal and HR-/HER2+ cells, their results significantly overlapped with the results of all three RNAi screens. Further statistical and experimental tests, including both gene silencing in cells and analysis of alternate datasets, narrowed down the field of possible master regulators, eventually converging on STAT3 as the only master regulator that was found in all HR-/HER2+ cells.
Mapping the HR-/HER2+ pathway
This finding was particularly interesting because it is consistent with a growing body of evidence suggesting that STAT3 is an important regulator of cancer cell homeostasis. Subsequent tests also showed that the activation of STAT3 was unique to HR- cells that had undergone transformation to cancer by the HER2 oncogene. The authors then pushed this finding farther, performing additional experiments and computational analyses in an effort to discern the entire pathway through which STAT3 drives the HR-/HER2+ subtype.
A schematic drawing of the signaling pathway responsible for HR-/HER2+ breast cancer and of how FDA-approved drugs could interrupt this program. (Source: Silva and Califano Labs)
Their investigation revealed a surprising and intriguing overlap between this cancer-driving pathway and other pathways known to be important in the immune response. They discovered that the well-known JAK/STAT pathway was uniquely activated in HR-/HER2+ cancer cells, and that there was a dramatic up-regulation of interleukin 6 (IL-6), an immune system signaling molecule that activates STAT3. In addition, gene expression profiling of cells that model HR-/HER2+ cells also showed a corresponding overexpression of S100A8 and S100A9, a pair of proteins that are known to be active in the immune system and in solid tumors associated with poor prognosis. Analysis of HR-/HER2+ cells in the METABRIC dataset, along with gene silencing experiments, confirmed the importance of these proteins to the function of STAT3.
Taken together, these findings offer a detailed picture of the program necessary to maintain HR-/HER2+ cells. The paper explains that secretion of IL-6 stimulates activation of STAT3 through the well-known JAK pathway. Once it is turned on, STAT3 induces upregulation of S100A8/9, which feeds other programs inside the cell known to be important in cancer.
“A key message emerging from this work,” Dr. Califano explains, “is that the regulatory logic of the cell can implement cancer-causing mechanisms that, albeit triggered by a mutation, become completely independent and self-sustained. This paper describes an autocrine loop involving IL-6 that is one example, and provides an additional rationale for why many tumors relapse following oncogene-targeted therapy.”
Testing drugs against HR-/HER2+ breast cancer
This overlap between HR-/HER2+ breast cancer and the immune system opens up some exciting therapeutic opportunities, particularly because previous research has already produced FDA-approved drugs that interfere with the function of this specific pathway. Although these drugs were initially approved for the treatment of other diseases, this finding offers tantalizing hope that they could quickly be repurposed as breast cancer therapeutics.
One existing drug that holds great potential is an antibody called tocilizumab, which blocks the effects of IL-6 and is approved for the treatment of rheumatoid arthritis. Another called ruxolitinib inhibits JAK1/2 and was recently approved for treatment of myelofibrosis, a bone marrow disorder that disrupts the production of normal blood cells. Ongoing research is also investigating tasquinimod — known to be capable of binding to S100A9 — for the treatment of solid tumors such as prostate cancer.
A clinical trial is now evaluating the effects of a combination therapy in patients with metastatic HER2-positive breast cancer.
Once the authors had characterized the pathway responsible for HR-/HER2+ breast cancer, they tested these three drugs in their disease model. As they expected, growth of HR-/HER2+ was significantly inhibited when the drugs were used alone and in combinations, and the drugs had little effect on normal cells without HER2 mutations. In a xenograft in which cancer cells from their model were implanted into mice, the investigators also saw a dramatic reduction in tumor growth.
These results have been so convincing that a multicenter phase I/II clinical trial is now underway under the direction of Kevin Kalinsky and Matthew Maurer, both physicians at Columbia University Medical Center. The trial is evaluating the safety and efficacy of a combination therapy including both ruxolitinib and trastuzumab in patients with metastatic HER2-positive breast cancer that has progressed even during treatment with standard HER2-targeted therapies. The trial is currently recruiting patients. (For more information about this trial, go to Clinicaltrials.gov.)
“If successful,” Dr. Silva remarks, “this new targeted therapy will represent a non-chemotherapy-containing anticancer strategy for HER2-positive breast cancers, and could also be applied to other HER2-positive human cancers.” The investigators will be watching the clinical trial closely to see if the hypothesis proves true.
— Chris Williams
Related publication
Rodriguez-Barrueco R, Yu J, Saucedo-Cuevas LP, Olivan M, Llobet-Navas D, Putcha P, Castro V, Murga-Penas EM, Collazo-Lorduy A, Castillo-Martin M, Alvarez M, Cordon-Cardo C, Kalinsky K, Maurer M, Califano A, Silva JM. Inhibition of the autocrine IL-6-JAK2-STAT3-calprotectin axis as targeted therapy for HR-/HER2+ breast cancers. Genes Dev. 2015 Aug 1;29(15):1631-48.