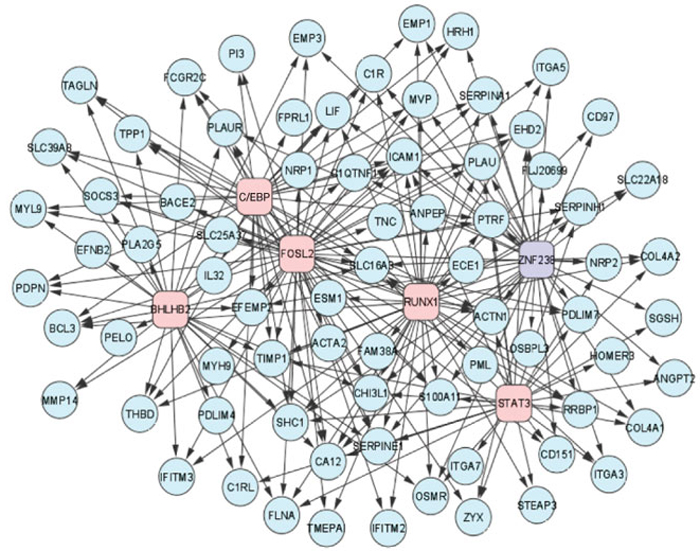
The mesenchymal signature of high-grade gliomas is controlled by six transcription factors. TFs involved in activation of MGES targets are shown in pink, those involved in repression are in purple.
High-grade gliomas, such as glioblastoma, are incurable partly because the tumor cells are widely disseminated throughout the brain. This capacity for invasive growth has been associated with the expression of genes more commonly transcribed in mesenchymal cells. In work published in the journal Nature, Antonio Iavarone, Andrea Califano, and colleagues have identified a small transcription factor network that is responsible for the mesenchymal behavior of glioma cells.
The authors applied a specific algorithm designed to infer causal transcription factor-target interactions to gene expression profiles from 176 samples of high-grade gliomas. They analyzed the resulting interactome with a new algorithm that enabled them to evaluate the transcription factor network in terms of a previously identified mesenchymal gene expression signature from high-grade gliomas. This identified the 53 transcription factors that are associated with regulating mesenchymal gene expression. Further analyses identified signal transducer and activator of transcription 3 (STAT3) and CAATT/enhancer binding protein-β (CEBPβ) as potential master regulators that control the expression of a substantial proportion of these mesenchymal genes. The authors conclude that systems biology approaches can be used to identify master transcription factors that are involved in malignant transformation, and such approaches could be applied to help dissect the complexity of other tumour phenotypes.