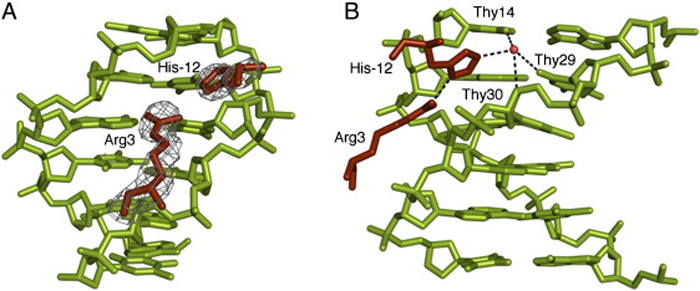
Minor Groove Insertion of Scr Residues His−12 and Arg3 in fkh250. (A) Electron densities for Arg3 and His−12 in the fkh250 complex. (B) Details of the His−12–Arg3 interaction and water-mediated interactions with Thy14, Thy29, and Thy30 of fkh250.
Although the basic structure of the double helix has been known since the classic work of Watson and Crick. it has become increasingly clear that the helix is not regular and that its shape depends on nucleotide sequence. In two recent papers in Cell and Nature, Barry Honig, Richard Mann and their colleagues in C2B2 and the Department of Biochemistry and Molecular Biophysics have shown that sequence-dependent variations in the helix shape allow DNA-binding proteins to recognize their specific binding sites.
This discovery was based initially on studies of Hox proteins that play a role in determining the anterior/posterior axis of embryos. Different Hox proteins must bind their various DNA targets with high specificity, and it was unclear how this was achieved. The researchers found that Hox proteins were able to recognize the width of the minor groove through the insertion of arginines in sites where the groove was narrow (Cell, 131:530, 2007). The newest findings, published in (Nature 461:1248, 2009), establish the generality of this mechanism and explain it physical origins. Specifically, short AT rich regions have an intrinsic tendency to narrow grooves and this in turn enhances the negative electrical potential of the DNA in this region, thus attracting positively charged arginines on protein surfaces. These findings are expected to have major impact on our ability to predict the DNA targets of different transcription factors.