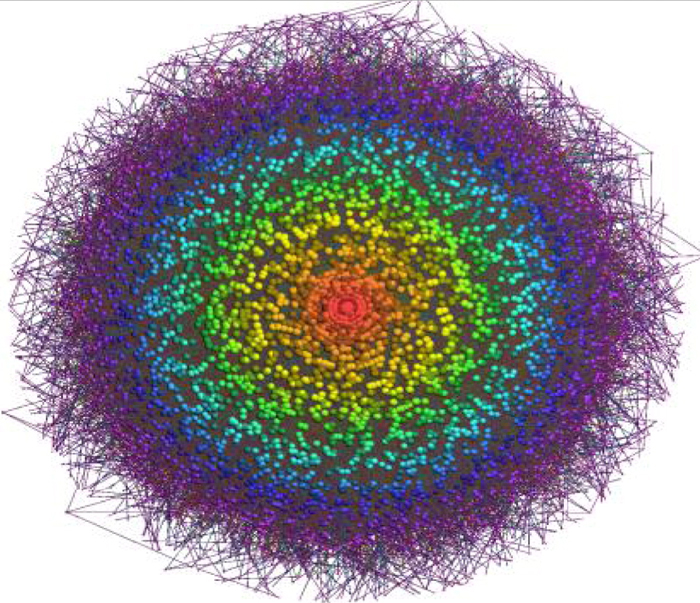
Genome-wide inference of sponge modulators identified a miR-program mediated post-transcriptional regulatory (mPR) network including ~248,000 interactions.
For decades, scientists have thought that the primary role of messenger RNA (mRNA) is to shuttle information from the DNA to the ribosomes, the sites of protein synthesis. However, new studies now suggest that the mRNA of one gene can control, and be controlled by, the mRNA of other genes via a large pool of microRNA molecules, with dozens to hundreds of genes working together in complex self-regulating sub-networks.
In work published in the journal Cell, Andrea Califano, José Silva, and colleagues analyzed gene expression data in glioblastoma in combination with matched microRNA profiles to uncover a posttranscriptional regulation layer of surprising magnitude, comprising more than 248,000 microRNA (miR)-mediated interactions. These include ∼7,000 genes whose transcripts act as miR “sponges.” When two genes share a set of microRNA regulators, changes in expression of one gene affects the other. If, for instance, one of those genes is highly expressed, the increase in its mRNA molecules will “sponge up” more of the available microRNAs. As a result, fewer microRNA molecules will be available to bind and repress the other gene’s mRNAs, leading to a corresponding increase in expression.
Although such an effect had been previously elucidated, the range and relevance of this kind of interaction had not been characterized.