News
Columbia Awarded NCI Center for Cancer Systems Biology
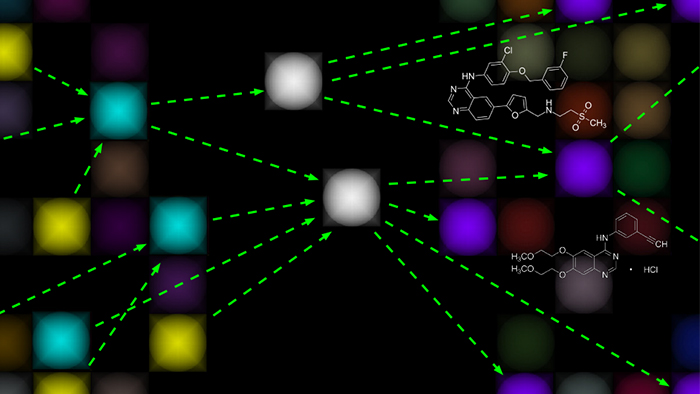
In this rendering, master regulators of tumor homeostasis (white) integrate upstream genetic and epigenetic events (yellow) and regulate downstream genes (purple) responsible for implementing cancer programs such as proliferation and migration. CaST aims to develop systematic methods for identifying drugs capable of disrupting master regulator activity.
The Columbia University Department of Systems Biology has been named one of four inaugural centers in the National Cancer Institute’s (NCI) new Cancer Systems Biology Consortium. This five-year grant will support the creation of the Center for Cancer Systems Therapeutics (CaST), a collaborative research center that will investigate the general principles and functional mechanisms that enable malignant tumors to grow, evade treatment, induce disease progression, and develop drug resistance. Using this knowledge, the Center aims to identify new cancer treatments that target master regulators of tumor homeostasis.
CaST will build on previous accomplishments in the Department of Systems Biology and its Center for Multiscale Analysis of Genomic and Cellular Networks (MAGNet), which developed several key systems biology methods for characterizing the complex molecular machinery underlying cancer. At the same time, however, the new center constitutes a step forward, as it aims to move beyond a static understanding of cancer biology toward a time-dependent framework that can account for the dynamic, ever-changing nature of the disease. This more nuanced understanding could eventually enable scientists to better predict how individual tumors will change over time and in response to treatment.
Investigating cancer in this way means sidestepping the “oncogene addiction” paradigm, which has focused on identifying and targeting individual genetic alterations that are critical for the survival of cancer cells. Although this approach has led to some notable successes (for example, imatinib, erlotinib, and herceptin), a small minority of tumors seen in the clinic manifest actionable mutations. Moreover, targeted inhibitors have by and large shown limited long-term effectiveness, typically failing as tumors relapse and become drug resistant. For this reason, identifying new categories of druggable targets and finding better ways to manage tumors as they evolve constitute important challenges for the future of cancer research.
Instead of focusing exclusively on actionable mutations, CaST will concentrate on the regulatory machinery that enables established tumors to survive. Driving its research is the hypothesis that cancer cells achieve and retain their malignant state by hijacking the regulatory programs responsible for normal cell development. Just as normal cells differentiate into specific cell types, tumor cells undergo a process of canalization in which molecular modules (tumor checkpoints) and specific proteins within these modules (master regulators) act like funnels, channeling the effects of upstream genetic alterations into cellular behaviors that are typical of cancer.
Such proteins, which appear consistently across many tumors, are typically not themselves the result of mutated genes. Nevertheless, they become essential for maintaining “tumor homeostasis” — the ability of cancer cells to remain in a stable state and grow. Previous work at Columbia has shown that master regulators are a type of “Achilles heel” for tumors, making them attractive targets for disrupting tumor homeostasis and cancer-related cell state transitions.
An additional component of CaST’s efforts will be to address the question of how the heterogeneity seen in tumors affects tumor homeostasis and canalization. Heterogeneity can manifest itself in the genetic diversity among the cells that make up an individual tumor, in the interactions between tumors and the microenvironments in which they grow, and in the molecular reprogramming that cells undergo over the course of disease. When subjected to anticancer drugs, this heterogeneity within a single tumor goes through a process of evolutionary selection, with different populations of cells reacting in different ways based on how their regulatory machinery processes the attack. The results of such selection pressures are often the downfall of existing therapies, as a portion of the tumor evades treatment and continues to grow.
Commonly used methods for profiling tumor tissue, including bulk sampling and sequencing techniques that generate an average profile of a tumor, have been incapable of distinguishing this fine-grained structure. For this reason, CaST will develop and test new methods for differentiating diverse cell populations within individual tumors. These tools will include new high-throughput single-cell sequencing technologies as well as patient-derived xenografts, in which human tumor tissue is implanted in mice to study how it grows and responds to different therapeutic approaches. Testing of these experimental methods will be accompanied by the development of new computational tools for analyzing the data that they and other methods generate.
Identifying tumor checkpoints and master regulators of specific tumor states, the scientists propose, could dramatically simplify the problem of prioritizing drugs and drug combinations against cancer. Using new high-throughput drug screening technologies developed in the Department of Systems Biology, the investigators plan to identify compounds whose mechanism of action can disrupt specific master regulator proteins responsible for tumor homeostasis and state transitions. Strategies for controlling cancer could then potentially include inducing cancer cell death, preventing progression to a malignant tumor stage, or reprogramming tumor cells so that they regain drug sensitivity.
CaST investigators will also address the dynamic nature of cancer by using RNAi and CRISPR screens to target master regulators, enabling them to explore how tumors adapt to pharmacological and genomic perturbations over long time spans. The resulting models, they hope, should provide a clearer picture of the “escape routes” that enable tumors to survive treatment.

Faculty involved in CaST include (from top left): Cory Abate-Shen, Dimitris Anastassiou, Andrea Califano, Aris Floratos, Barry Honig, Andrew Kung (MSKCC), Kenneth Olive, Itsik Pe'er, Raul Rabadan, Peter Sims, Nicholas Tatonetti, Dennis Vitkup, and Harris Wang.
“We are very excited to have the opportunity to explore how systems biology approaches could help address some of the most critical questions facing precision cancer medicine today,” says Andrea Califano, chair of the Department of Systems Biology and a co-principal investigator for the center. “By assembling this multidisciplinary team of very dynamic scientists, we think CaST should be in a unique position to push the field forward.”
In addition to Dr. Califano, Barry Honig, director of the Center for Computational Biology and Bioinformatics, will also lead CaST as co-PI. Other faculty and investigators involved in the project include Cory Abate-Shen, Dimitris Anastassiou, Aris Floratos, Charles Karan, Andrew Kung, Diana Murray, Kenneth Olive, Itsik Pe’er, Raul Rabadan, Peter Sims, Nicholas Tatonetti, Dennis Vitkup, and Harris Wang.
The award is particularly notable for the Department of Systems Biology because it now has grants from all three of the NCI’s programs supporting research centers in this field. Other Department of Systems Biology national centers are funded through the Cancer Target Discovery & Development Program and the Physical Science-Oncology Program.
For more information about the Center for Cancer Systems Therapeutics, visit the CaST website.
— Chris Williams