News
New Method for Identifying Genetic Alterations that Modulate Gene Expression
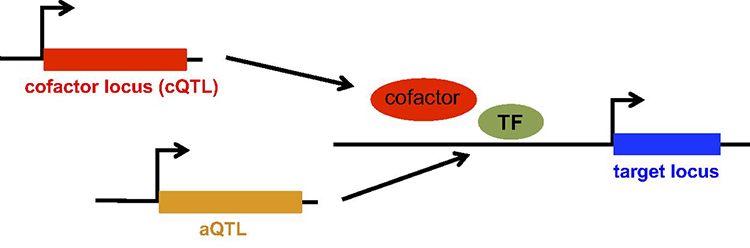
Cofactors work with transcription factors (TFs) to enable efficient transcription of a TF's target gene. The Bussemaker Lab showed that genetic alterations in the cofactor gene (cQTLs) change the nature of this interaction, affecting the connectivity between the TF and its target gene. This, combined with other factors called aQTLs that affect the availability of the TF in the nucleus, can lead to downstream changes in gene expression.
When different people receive the same drug, they often respond to it in different ways — what is highly effective in one patient can often have no benefit or even cause dangerous side effects in another. From the perspective of systems biology, this is because variants in a person’s genetic code lead to differences in the networks of genes, RNA, transcription factors (TFs), and other proteins that implement the drug’s effects inside the cell. These multilayered networks are much too complex to observe directly, and so systems biologists have been developing computational methods to infer how subtle differences in the genome sequence produce these effects. Ultimately, the hope is that this knowledge could improve scientists’ ability to identify drugs that would be most effective in specific patients, an approach called precision medicine.
In a paper published in the Proceedings of the National Academy of Sciences, a team of Columbia University researchers led by Harmen Bussemaker proposes a novel approach for discovering some critical components of this molecular machinery. Using statistical methods to analyze biological data in a new way, the researchers identified genetic alterations they call connectivity quantitative trait loci (cQTLs), a class of variants in transcription cofactors that affect the connections between specific TFs and their gene targets.
The work grew from the Bussemaker Lab’s longstanding interest in understanding how transcription factors regulate the transcription of genes into mRNA. Transcription factors are proteins that promote or repress expression of particular genes by binding to specific sequences of DNA. Genetic alterations in a TF’s binding sites in genomic DNA can alter its binding and lead to changes in gene expression. In some cases this can lead to disease. For this reason, many scientists are searching for such alterations in the genome, which they call expression quantitative trait loci (eQTLs).

Harmen Bussemaker
In 2010 Bussemaker proposed the existence of a new type of QTL. These so-called transcription factor activity QTLs (aQTLs) do not affect the TF binding sites themselves, but rather influence how much of a TF protein in the cell is present in the nucleus, where the genomic DNA resides. More recently, in collaboration with researchers at the Netherlands Cancer Institute, his lab used an extension of the aQTL approach called locus expression signature analysis (LESA) to analyze how viral insertions cause cancer in mice by affecting the protein-level activity of specific TFs. They also extended the aQTL approach to post-transcriptional networks that regulate mRNA stability.
The new PNAS paper extends this line of thought further by considering the fact that a transcription factor does not work alone to control gene expression. Instead, it relies on interactions with cofactor gene products that enhance the strength of its binding to its target genes, or its ability to interact with proteins that help it activate or repress its gene targets. Bussemaker hypothesized that genetic alterations in those cofactors could change their interaction with their partner transcription factors, altering the strength of TF binding and thereby influencing downstream gene expression. This could in turn, for example, change the way in which the cell processes an incoming signal from a drug. The team calls these alterations cQTLs.
The algorithm for identifying cQTLs grew from a PhD thesis by Mina Fazlollahi, a former graduate student in the Bussemaker Lab who is now a postdoctoral scientist at Mount Sinai School of Medicine. Running the algorithm requires data representing genome sequence, gene expression levels, and prior information about transcription factor binding preferences. It then integrates and analyzes the data in several steps using statistical approaches described in the paper.
The scientists tested their method in two yeast strains, sampling approximately 100 known transcription factors and then focusing on seven whose functional connectivity with their target genes seemed to be affected by genetic alterations. One of these, called Ste12p, was previously known to be an activator of the mating response pathway in yeast in the presence of a small molecule called α-pheromone. They applied their algorithm to find cQTLs related to Ste12p, and then used existing protein-protein interaction data to identify protein products within these regions that are physically capable of interacting with the transcription factor as cofactors.
One of the regions they pinpointed included a gene called DIG2, which had previously been shown to be a regulator of Ste12p activity. To determine whether the naturally occurring genetic alteration in the DIG2 protein was indeed a cQTL for Ste12p, they engineered a strain that was previously susceptible to α-pheromone so that it now differed by a single nucleotide in DIG2. Stimulating the strain with the small molecule led to a dramatic drop in Ste12p-mediated gene expression. Conversely, they engineered another strain that was previously not responsive to α-pheromone so that it contained the active nucleotide sequence in the predicted cQTL, and saw that it subsequently displayed the Ste12p expression activity normally seen in the mating response. Being able to control gene expression in this way, along with other findings described in the paper, confirmed the ability of the researchers’ algorithm to identify cQTLs accurately. The experiments were performed under the supervision of Helen Causton, an Assistant Professor in the Department of Pathology and Cell Biology at Columbia University Medical Center
Although their method was applied to a humble yeast system, the authors indicate that it should offer a way to identify cQTLs in any organism for which the necessary data are available. “So far, I like to think of this as personalized medicine for yeast,” Bussemaker remarks. “But with the increasing availability of genotype and expression profile data in projects like the Genotype Tissue Expression (GTEx) project, and considering the recent arrival of high-quality in vitro protein-DNA interaction data, we think we should be able to apply these same methods to identify cQTLs in humans as well.” Refining their approach and investigating this newly characterized layer of gene regulation in mammalian systems is an ongoing project in the Bussemaker Lab.
— Chris Williams
Related publications
Fazlollahi M, Muroff I, Lee E, Causton HC, Bussemaker HJ. Identifying genetic modulators of the connectivity between transcription factors and their transcriptional targets. Proc Natl Acad Sci U S A. 2016 Mar 10. [Epub ahead of print]
Lee E, Bussemaker HJ. Identifying the genetic determinants of transcription factor activity. Mol Syst Biol. 2010 Sep 21;6:412.
Lee E, de Ridder J, Kool J, Wessels LF, Bussemaker HJ. Identifying regulatory mechanisms underlying tumorigenesis using locus expression signature analysis. Proc Natl Acad Sci U S A. 2014 Apr 15;111(15):5747-52.
Fazlollahi M, Lee E, Muroff I, Lu XJ, Gomez-Alcala P, Causton HC, Bussemaker HJ. Harnessing natural sequence variation to dissect posttranscriptional regulatory networks in yeast. G3 (Bethesda). 2014 Jun 17;4(8):1539-53.