News
Researchers Identify RNA Binding Protein that Promotes Breast Cancer Metastasis
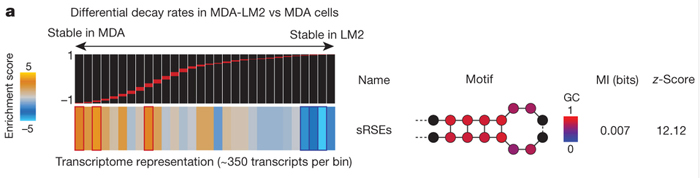
The presence of the structural RNA stability element (sSRE) family of mRNA elements distinguishes transcript stability in metastatic MDA-LM2 breast cancer cell lines from that seen in its parental MDA cell line. Each bin contains differential decay rate measurements for roughly 350 transcripts. From left (more stable in MDA) to right (more stable in MDA-LM2), sRSE-carrying transcripts were enriched among those destabilized in MDA-LM2 cells. The TEISER algorithm collectively depicts sSREs as a generic stem-loop with blue and red circles marking nucleotides with low and high GC content, respectively. Also included are mutual information (MI) values and their associated z-scores.
Gene expression analysis has become a widely used method for identifying interactions between genes within regulatory networks. If fluctuations in the expression levels of two genes consistently shift in parallel over time, the logic goes, it is reasonable to hypothesize that they are regulated by the same factors. However, such analyses have typically focused on steady-state gene expression, and have not accounted for modifications that messenger RNAs (mRNAs) can undergo during the time between their transcription from DNA and their translation into proteins. Researchers now understand that certain stem loop structures in mRNAs make it possible for proteins to bind to them, often causing RNA degradation and subsequently modulating protein synthesis. From the perspective of systems biology, this can have implications for the activity of entire regulatory networks, and recent studies have even suggested that aberrations in mRNA stability can play a role in disease initiation and progression.
In a new paper published in the journal Nature, Department of Systems Biology Professor Saeed Tavazoie and collaborators at the Rockefeller University describe a new computational and experimental approach for identifying post-transcriptional modifications and investigating their effects in biological systems. In a study of metastatic breast cancer, they determined that when the protein TARBP2 binds to a specific structural element in mRNA transcripts, it increases the likelihood that cancer cells will become invasive and spread. Interestingly, they also found that TARBP2 causes metastasis by binding transcripts of two genes — amyloid precursor protein (APP) and zinc finger protein 395 (ZNF395) — that have previously been implicated in Alzheimer’s disease and Huntington’s disease, respectively. Although the nature of this intersection between the regulatory networks underlying cancer and neurodegenerative diseases is unclear, the finding raises a tantalizing question about whether these very different disorders might be linked at some basic biological level.
The project was led by Hani Goodarzi, a former graduate student in the Tavazoie Lab who is now a postdoctoral scientist working with Sohail Tavazoie at the Rockefeller University. Sohail, who is also Saeed's brother, is an experimental scientist whose work has focused on mapping the post-transcriptional landscape of cancer metastasis. As a graduate student Goodarzi developed an algorithm called TEISER (Tool for Eliciting Informative Structural Elements in RNA), which systematically considers the entire range of small structural motifs that might be present in an RNA and computationally identifies those that correlate strongly with specific characteristics of RNA activity. In this new study, Goodarzi adapted TEISER to compare the decay rates of approximately 13,000 transcripts expressed in two closely related breast cancer cell lines, one of which is highly metastatic. The algorithm revealed a stem loop structure that was enriched in RNA transcripts that had a higher decay rate in highly metastatic cells relative to the genetically related non-metastatic variant cell-line. Experimental validation showed that this structure, which the researchers termed a structural RNA stability element (sSRE), was sufficient to suppress gene expression in the presence of an sSRE-binding factor. The researchers also showed that this effect was due to the stem loop structure, not the transcript’s nucleotide sequence.
“This computational structural motif discovery algorithm was able to find the structural RNA regulatory element through which transcript stability was differentially modulated,” Saeed Tavazoie explains. “Computation didn't just provide supportive analysis; it was critical in taking the large dataset and identifying this RNA stem loop structure.”
To identify the sRSE-binding protein that causes transcript degradation in metastatic breast cancer cells, the authors used a more conventional gene expression analysis to search for RNA binding proteins whose gene expression profiles closely match those of the sRSE-carrying transcripts. The analysis yielded three candidates, of which TARBP2 was the only one to show a clear effect on the expression of sRSE-containing transcripts in a series of gene knockout experiments in cell lines and in mice. Although TARBP2 had previously been known to play a role in microRNA processing, the discovery that the protein could bind stem loop structures in mRNAs was a new discovery, showing that this single protein has at least two distinct functions within the cell.
Follow-up experiments using HITS-CLIP, a genome-wide technology for mapping protein-RNA binding sites in vivo, clearly showed that TARBP2 binds to mRNAs at the stem loop structure that TEISER had predicted. The researchers also confirmed that TARBP2 binding destabilizes the sRSEs. Clinical association studies and knockdown experiments in living mice provided additional confirmation that the high expression of TARBP2 significantly raised the invasiveness of cancer cells.
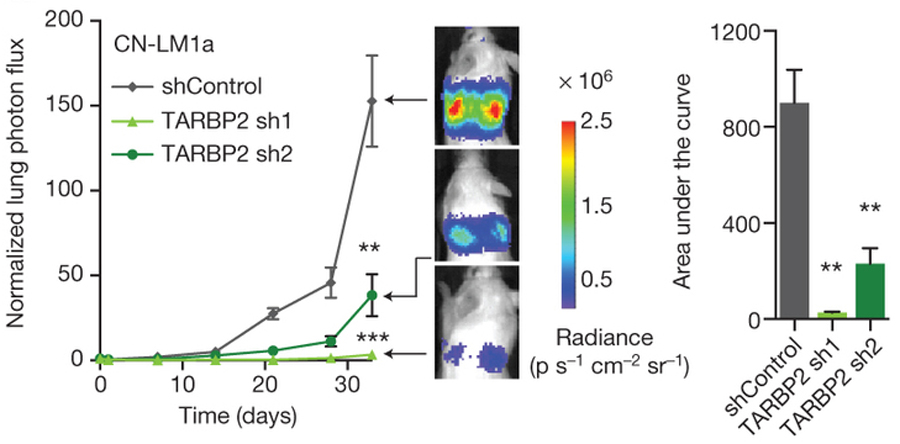
CN-LM1a cells expressing short hairpin RNAs (shRNAs) targeting TARBP2 (TARBP2 sh1 and sh2) or a control hairpin (shControl) show a dramatic drop in metastasis when compared with controls. The area under the curve was also calculated for each mouse (change in normalized lung photon flux times days elapsed).
Once TARBP2’s role in promoting metastasis was determined, the researchers sought to identify the specific genes whose transcripts contain the stem loop structure bound by the protein. By conducting an unbiased analysis of gene expression and stability profiles of cells in which TARBP2 was knocked out, they identified four transcripts that are directly bound by the protein in vivo and are destabilized and downregulated in highly metastatic cells. The team was surprised to find that the results included transcripts for APP, which is known to play an important role in Alzheimer's disease, and ZNF395, which has been implicated in Huntington’s disease. They also found that patients whose tumors produced more of these transcripts showed lower amounts of metastasis. These findings point to an inverse relationship between TARBP2’s ability to promote metastasis and levels of APP and ZNF395, suggesting that these two genes function as suppressors of metastasis in breast cancer cells.
What this intersection between the regulatory networks underlying breast cancer and neurodegenerative diseases might mean is still unclear, but it presents a fascinating problem. “We don’t know what’s going on,” Tavazoie says. “It could be purely coincidental, or it could hint at a deeper connection between the basic biology of metastasis and the basic processes that go awry in neurodegenerative disorders.”
Even with these open questions, however, this fruitful collaboration between computational and experimental scientists offers a powerful new approach for exploring the largely uncharted landscape of post-transcriptional modifications, whether due to protein-RNA binding or other phenomena such as alternative splicing and RNA localization that can also modulate protein production in both space and time. It may even open the door to developing therapeutic interventions that use small molecules or oligonucleotides to interfere with disease-driving interactions between proteins and mRNA.
— Chris Williams
Related publication
Goodarzi H, Zhang S, Buss CG, Fish L, Tavazoie S, Tavazoie SF. Metastasis-suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature. 2014 9 Jul.