News
Method for Determining Protein Function Opens Opportunities for Precision Cancer Medicine
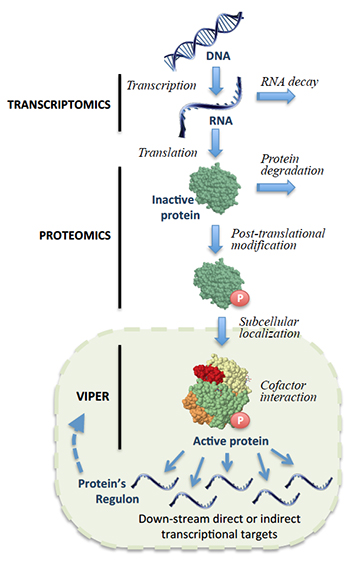
Following gene transcription and translation, a protein can undergo a variety of modifications that affect its activity. By analyzing downstream gene expression patterns in single tumors, VIPER can account for these changes to identify proteins that are critical to cancer cell survival.
In a paper just published in Nature Genetics, the laboratory of Andrea Califano introduces what it describes as the first method capable of analyzing a single tumor biopsy to systematically identify proteins that drive cancerous activity in individual patients. Based on knowledge gained by modeling networks of molecular interactions in the cell, their computational algorithm, called VIPER (Virtual Inference of Protein activity by Enriched Regulon analysis), offers a unique new strategy for understanding how cancer cells survive and for identifying personalized cancer therapeutics.
Developed by Mariano Alvarez as a research scientist in the Califano laboratory, VIPER has become one of the cornerstones of Columbia University’s precision medicine initiative. Its effectiveness in cancer diagnosis and treatment planning is currently being tested in a series of N-of-1 clinical trials, which analyze the unique molecular characteristics of individual patients’ tumors to identify drugs and drug combinations that will be most effective for them. If successful, it could soon become an important component of cancer care at Columbia University Medical Center.
According to Dr. Califano, “VIPER makes it possible to find actionable proteins in 100% of cancer patients, independent of their genetic mutations. It also enables us to track tumors as they progress or relapse to determine the most appropriate therapeutic approach at different points in the evolution of disease. So far, this method is looking extremely promising, and we are excited about its potential benefits in finding novel therapeutic strategies to treat cancer patients.”
From oncogenes to oncoproteins
At its core, cancer is a constellation of diseases that arise when normal protein activity in cells goes awry, causing them to grow and spread uncontrollably. Because proteins are the products of genes, cancer biologists have for many years hypothesized that tumor cells become addicted to the mutated oncogenes that are responsible for the initial tumor growth. By identifying and targeting the proteins harboring these mutations, the reasoning goes, it should be possible to design personalized therapies that could halt cancer in its steps.
Research in this direction has led to some important successes, including development of the blockbuster drugs imatinib, erlotinib, and herceptin. In general, however, it has thus far improved treatment for only a small minority of tumors. At the opening plenary session of the 2016 meeting of the American Association for Cancer Research (AACR), Elaine Mardis (McDonnell Genome Institute at Washington University) reported that only 11% of cancer patients treated with this strategy see an increased disease-free survival, leaving nearly 90% without access to viable targeted therapies. This suggests a need for other complementary approaches.
In part, this longstanding focus on genes as a proxy for protein activity has been a consequence of the strengths and limitations of available technologies. Next-generation DNA sequencing is consistently reproducible, a factor that is essential for clinical applications. However, gene sequence alone cannot reveal whether the corresponding protein is actually aberrantly activated. This is because proteins operate in cooperation, collectively forming complex interaction networks that influence and ultimately determine whether a mutation will affects a cell’s behavior, if at all.
Even when cancer-driving mutations have been identified, researchers have also faced an uphill battle in identifying durably effective therapeutics. One reason is that tumors typically develop drug resistance as they evolve. Another is that some of the most commonly recurring mutations block the activity of genes called tumor suppressors, such as TP53 and PTEN. Since drugs generally work by inhibiting proteins — not activating them — mutated tumor suppressor proteins do not provide good therapeutic targets.
From this perspective, being able to identify cancer-driving oncoproteins through direct measurement of protein activity across the course of disease would be more desirable. But although current technologies such as mass spectrometry can measure protein abundance, they are too expensive and complicated for use in clinical applications, and cannot systematically account for a wide range of factors — such as post-translational modifications or protein localization in specific parts of a cell — that affect protein activity. New methods for identifying the proteins responsible for driving cancer would therefore be a valuable addition in the fight against disease.
Virtual proteomics
The Califano Lab’s approach to identifying oncoproteins is based on past work revealing that although cancer cells can harbor an extremely heterogeneous repertoire of genetic alterations, these mutations enable them to misuse the complex network of molecular interactions that regulate their behavior in extremely similar ways. Even though many genetic mutations are present in tumor cells, Califano has shown that these mutations converge to and are integrated by specific proteins called master regulators, which activate the programs that are necessary to make a cancer cell. Importantly, these proteins are not themselves mutated, but are nevertheless essential for maintaining cells in their cancer-related state. Previous work has also indicated that these master regulators are relatively few and are conserved across a large subset of cancer patients. Identifying them and finding ways to target them could thus simplify the landscape of cancer dramatically, especially when compared to the myriad ways in which a tumor cell’s genome can be mutated.
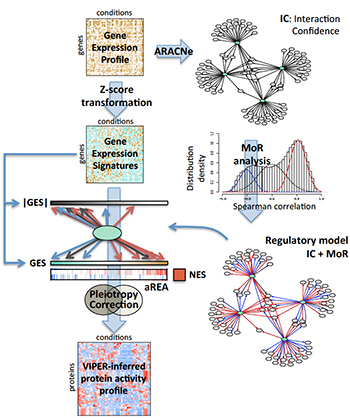
During a typical VIPER analysis, a regulatory network specific to the tumor being studied is assembled using ARACNe. Each interaction is then analyzed to determine whether target genes are activated or repressed (MoR), generating a tumor-specific model of how gene transcription is regulated. In parallel, genome-wide gene expression data is used to generate gene expression signatures for the sample. An algorithm called aREA then interprets the signatures, using the regulatory model to infer the relative activity of the proteins that regulate expression. In this way, gene expression profiles are transformed into regulatory protein activity profiles.
A previous algorithm that Califano developed, called MARINa, first made it possible to investigate and validate this hypothesis, and has been used to identify master regulators for breast and prostate cancer, glioma, lymphoma, leukemia, and many other cancer subtypes. However, MARINa requires multiple samples representing the same tumor subtype to identify master regulators. Their new algorithm, called VIPER, offers a statistically robust method for achieving the same objective using a single patient tumor sample.
VIPER is based on a very simple concept. Rather than measuring the activity of a protein directly — an extremely difficult task — it infers activity based on the expression of the genes the protein regulates. In the method described in their paper, the Califano Lab first uses ARACNe, an algorithm that has been broadly adopted and validated by the research community, to identify targets of all proteins in a specific tumor type. By applying a novel statistical framework for analyzing gene expression data generated from a single tumor, VIPER then determines the activity of all cancer-relevant proteins, identifying those that are abnormally activated in a specific tumor.
“It’s like detective work to determine which of two crime families was the mastermind behind a murder,” Califano explains. “First you build a map of the two organizations and then look for fingerprints or eyewitness accounts of who was at the crime scene. If you identify someone who is a part of one of the two organizations, you can quickly figure out the head of the organization who gave the order. In a similar way, we can understand protein activity by observing expression changes in the genes they regulate.”
Importantly, the researchers report that because VIPER measures each protein’s activity based on the expression of hundreds of genes, their measurements are highly reproducible and thus appropriate for clinical utilization. This is the case even though individual gene expression measurements may not be sufficiently reproducible. This feature makes it possible to investigate protein activity using formalin-fixed, paraffin-embedded (FFPE) tissue samples, which are clinically more common than fresh tissue samples but are typically degraded in ways that have in the past made them difficult to analyze.
In addition, drugs are already known to be capable of targeting many of the specific proteins that VIPER identifies. And because RNA sequencing (RNASeq) is less expensive than genome sequencing — and VIPER requires just a single tissue sample — this approach is a tenth of the cost of genetic sequencing. This makes it feasible to use the algorithm repeatedly during the course of a patient’s cancer treatment. For example, if a tumor stops responding to a particular therapy, a new VIPER analysis could be performed to determine how it has evolved and which new druggable proteins are now essential for its survival.
Focus on networks simplifies cancer diagnosis
As the paper reports, the Califano Lab performed a number of studies to validate VIPER’s effectiveness. In one they analyzed 173 basal breast carcinomas recorded in the Cancer Genome Atlas (TCGA). Even though these samples were ostensibly the same type of cancer, the investigators found that their gene expression patterns were wildly different from sample to sample, making it very difficult to predict a therapy that might work for all patients from expression alone. However, when they used the network-based approach that VIPER offers, they discovered that essential cancer-driving proteins were consistently present across all samples. Such findings are exciting in light of other work in the Califano Lab that has indicated that these proteins — which are different from the typical oncogenes like BRAF, EGFR, and ERBB2 — could be important tumor checkpoints across many cancer types.
“This makes it possible to use a more universal treatment for all cancers in this subtype,” Califano explains. “If there are drugs that can target those proteins, you don’t need to figure out how each cancer is different at the genetic level.”
In the end, the scientists used VIPER to investigate more than 10,000 tumor samples representing 14 different malignancies from the TCGA repository. Their findings suggest that VIPER can identify dysregulation in cancer cells that results both from mutations and from proteins whose abnormal activity did not arise from mutations in their corresponding genes but gained their cancer-driving ability because of other alterations. This suggests that currently available drugs could be used effectively in a substantial subset of patients.
VIPER powers a new approach to precision medicine
As VIPER has developed, it is quickly becoming an important tool in Columbia’s precision medicine initiative. This University-wide effort is investigating new approaches and technologies to improve personalized treatment for cancer and other diseases based on molecular characteristics in individual patients. In this context, the new approach described in the paper complements and extends other key avenues being pursued, including immunotherapy and genomic medicine.
Systems biology-based methods developed in the Califano Lab, including most importantly VIPER, have become the basis of a series of N-of-1 cancer clinical trials at Columbia University Medical Center. Working with clinical researchers in the Herbert Irving Comprehensive Cancer Center, scientists in the Califano Lab use VIPER to analyze tumor samples from individual patients, identify proteins that are driving cancerous activity, and connect them to existing FDA-approved and investigative drugs that are already known to be able to target them. Although directly providing treatment is beyond the scope of the trials, findings resulting from these studies have already enabled Columbia oncologists to recommend therapies that have extended survival and improved quality of life in patients.
As they continue testing their new approach to cancer diagnosis and treatment planning, Califano is cautiously optimistic for future applications of VIPER, saying, “If the preliminary findings in the N-of-1 trials are further confirmed, this approach could become an important tool.”
— Chris Williams
Related publication
Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, Califano A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016 Jun 20.