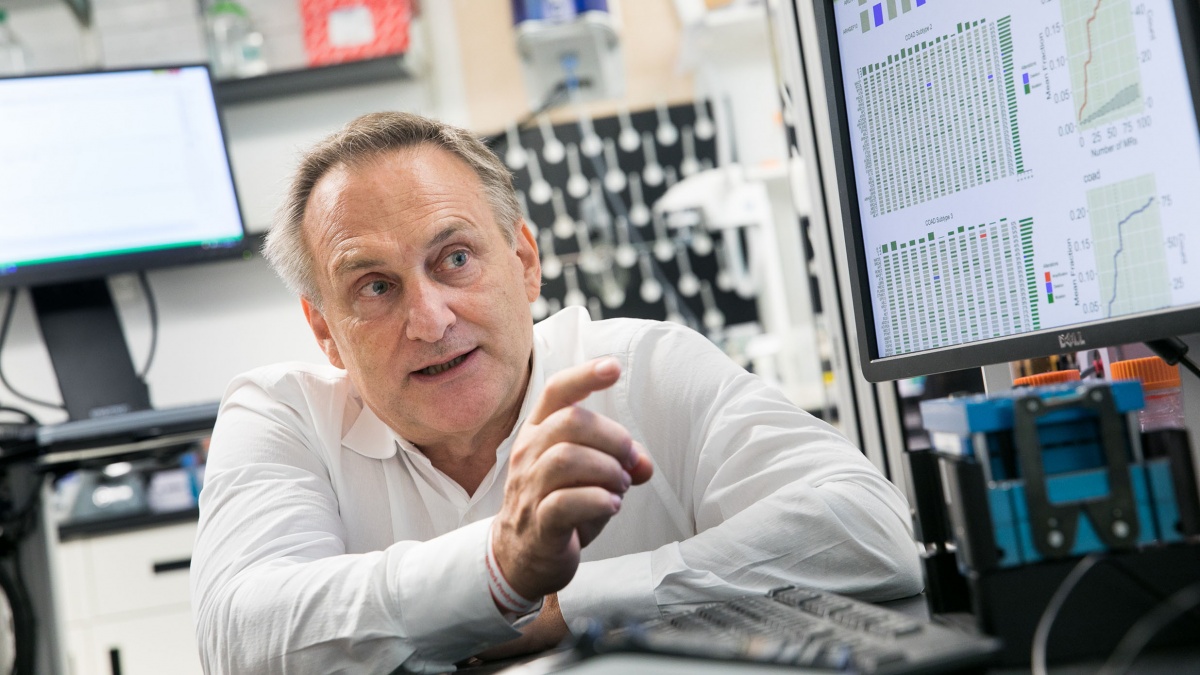
In recognition of Dr. Andrea Califano's recent Ruth Leff Siegel Award , an annual prize that honors and supports an investigator who has made outstanding contributions to our understanding of pancreatic cancer, Let's Win! Pancreatic Cancer has published the following feature article spotlighting his innovative approach to cancer research.

Dr. Andrea Califano, 2019 recipient of annual Ruth Leff Sigel Award for pancreatic cancer research. (Photo: Jörg Meyer/Columbia Magazine)
If you look at our basic biology, humans are big, cumbersome living organisms with a lot of moving parts.
For most of our lives, the cellular machinery that keeps us functioning goes off without a hitch. Starting at conception, cells have been growing and dividing, structuring themselves in a highly organized fashion. Liver cells know their job. And brain and spinal cord cells know their jobs, too.
Cancer is also a living organism. After all, it grows and evolves just like healthy cells. But cancer cells are cheats, ignoring the rules that other healthy cells play by. They mutate and divide uncontrollably, finding ways to evade our immune systems, which try to keep these invaders in check. To complicate matters, cancer cells are what scientists call heterogeneous. That means that even in the same malignant tumor there can be a variety of mutations, which is one reason why cancer treatment often fails. Drugs simply can’t target all of those mutations.







 Download PDF
Download PDF