News
A First Method for Global Probabilistic Annotations of Metabolic Networks
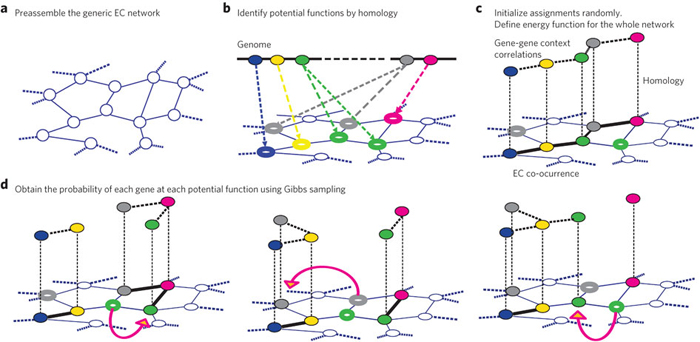
An overview of the GLOBUS algorithm.
A Columbia University team led by professor Dennis Vitkup and PhD student German Plata of the Center for Computational Biology and Bioinformatics has developed a novel genome-wide framework for making probabilistic annotations of metabolic networks. Their approach, called Global Biochemical Reconstruction Using Sampling (GLOBUS), combines information about sequence homology with context-specific information including phylogeny, gene clustering, and mRNA co-expression to predict the probability of biochemical interactions between specific genes. By integrating these different categories of information using a principled probabilistic framework, this approach overcomes limitations of considering only one functional category or one gene at a time, providing a global and accurate prediction of metabolic networks.
In a paper published in Nature Chemical Biology, the scientists write, "Currently, most publicly available biochemical databases do not provide quantitative probabilities or confidence measures for existing annotations. This makes it hard for the users of these valuable resources to distinguish between confident assignments and mere guesses... The GLOBUS approach, which is based on statistical sampling of possible biochemical assignments, provides a principled framework for such global probabilistic annotations. The method assigns annotation probabilities to each gene and suggests likely alternative functions."
To test the accuracy of GLOBUS, the Columbia researchers used it to generate genome-wide predictions about the metabolic networks in the bacteria Bacillus subtilis and Staphylococcus aureus, and selected three predictions for experimental validation. Subsequent laboratory experiments by their collaborators at ETH Zurich confirmed the predicted functions of these genes. Among the predictions is an important pathway of genes responsible for spore formation in B. subtilis.
The researchers suggest that data from GLOBUS can be easily combined with metabolomic, proteomic, and fluxomic data being generated by experimental methods. By identifying complementary patterns that appear across these different data modalities, scientists will be able to make increasingly confident predictions about the function of genes involved in metabolism. The paper also suggests that GLOBUS could be particularly important for understanding metabolic networks in less-studied organisms.
In addition to the paper, see a News and Views commentary in Nature Chemical Biology about this novel approach.
— Chris Williams
Related publication
Plata G, Fuhrer T, Hsiao TL, Sauer U, Vitkup D. Global probabilistic annotation of metabolic networks enables enzyme discovery. Nat Chem Biol. 2012 Oct;8(10):848-54.