News
Columbia Researchers Will Use Master Regulators to Reclassify Cancer Subtypes

Andrea Califano and Aris Floratos will lead an effort to reclassify tumors catalogued in TCGA according to their master regulators.
Andrea Califano and Aris Floratos, faculty members in the Columbia University Department of Systems Biology, have received a two-year, $624,236 subcontract to develop a new classification system of cancer subtypes. The agreement was awarded through a subcontract from Leidos Biomedical Research, Inc., which operates the Frederick National Laboratory for Cancer Research for the federal government.
By performing an integrative analysis of genomic data from the Cancer Genome Atlas (TCGA) and proteomic data from the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC), the researchers plan to recategorize tumors collected in TCGA based on the master regulator genes that determine their state. This is in contrast to other approaches based on expression of genes that reflect tissue lineage and proliferative processes. In addition, the team will link the genetics of each tumor sample to the specific master regulators that determine its state using a recently published novel algorithm (DIGGIT). Ultimately, the project aims to provide a more useful catalog of pan-cancer subtypes that could help to identify biomarkers and therapeutic targets for specific kinds of tumors, and ultimately provide a resource to guide the next generation of precision medicine.
“We have to reevaluate the way in which we organize tumors within subtypes, using both gene expression data and mutational data,” says Dr. Califano. “Right now the common approach is to classify tumor types based on rather generic genes that are differentially expressed between subtypes. But most of these genes play no role in actually driving the disease. We want to shift the emphasis and classify tumors based on the genes that truly regulate tumor state and survival.”
Over the past 10 years, the Califano Lab has developed a suite of computational methods for modeling cell regulatory interaction networks (also called interactomes), demonstrating that interactomes of particular cancer subtypes become “rewired” in consistent and predictable ways. They have also repeatedly found that such networks allow systematic identification of genes called master regulators, which represent regulatory bottlenecks that are necessary and sufficient to establish and maintain tumor state. Such master regulators are infrequently mutated and thus evade detection by conventional mutational studies. Yet a number of them are essential for tumor survival and many occur in synthetic lethal pairs, where neither gene in isolation is essential for tumor survival but the pair is.
Under this new subcontract, the Califano Lab will apply this perspective to identify the master regulators of every tumor represented in TCGA, on a sample-by-sample basis. In addition to using TCGA data — which contains data about a tumor’s mutations, gene expression, and other genomic information — they will also make use of proteomic data from the CPTAC Data Portal, which contains mass spectrometry measurements related to protein identity, protein abundance, and post-translational modifications that can degrade a protein or change its regulatory activity. Incorporating such high-quality experimental data into this systems biology-based computational approach will make it possible to develop reliable models of how the various proteins in the network work together to drive disease.
Once the master regulators for all of the tumors in the database have been identified, tumor samples in the top 20 cancer types represented in TCGA will be reclassified, using a pan-cancer approach. Doing so, Califano anticipates, will reveal a limited repertoire of master regulators that ultimately drive a large faction of the tumors, many of which should be independent of traditional organ based tumor classification. Using the DIGGIT algorithm, they will also look upstream of master regulators, within regulatory networks, to identify the genomic and epigenomic alterations that determine their aberrant activity.
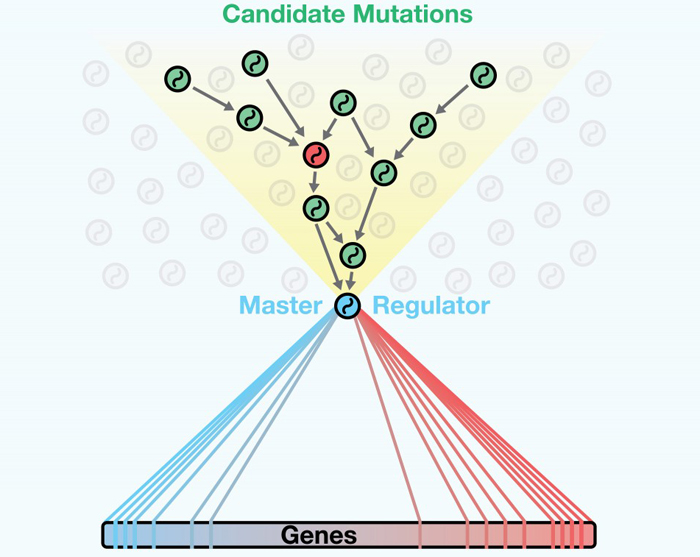
DIGGIT looks upstream of master regulators to distinguish driving mutations of cancer from other genes that might be mutated, but are not actually involved in causing and maintaining cancerous cell states.
Knowing the mutations and master regulators that drive these newly defined cancer subtypes will dramatically simplify the landscape for future cancer genome research. The project is just beginning, but based on initial data Califano anticipates that they might find that as few as 250 proteins, out of a genome containing more than 20,000 genes, may turn out to be responsible for driving a majority of tumors.
“Based on findings we published in a study of prostate cancer in 2013,” Califano says, “we think this limited number of master regulators will be essential in selecting a very useful panel of secreted protein biomarkers that could be monitored in the blood. Once they are identified, our hope is that a test could be developed that would look for those proteins or DNA transcripts in a blood test, and provide valuable information that could be used to guide personalized early cancer detection and treatment.”
As a final step, the Califano Lab will search for existing, FDA-approved drugs that have already been shown to target the mutations and master regulators that their analyses reveal. Applying their findings in this way could also yield practical insights that could be used to control or eliminate tumors. These will complement traditional genetic-based approaches that match small molecule inhibitors to mutated oncogenes.
To maximize the impact of the project’s findings, Dr. Floratos will oversee the integration of all data into geWorkbench, a web-based portal that provides easy access to the Columbia University Department of Systems Biology’s computational tools. In addition, the entire computational pipeline developed for the project, called Citrus, will be implemented as a cloud-based service on Google’s Compute Engine Infrastructure. This will enable cancer researchers anywhere in the world to access the data and regulatory models that the project generates, and to perform their own analyses as new cancer data become available.
— Chris Williams