News
Algorithm Identifies Genetic Driver of Mesenchymal Glioblastoma
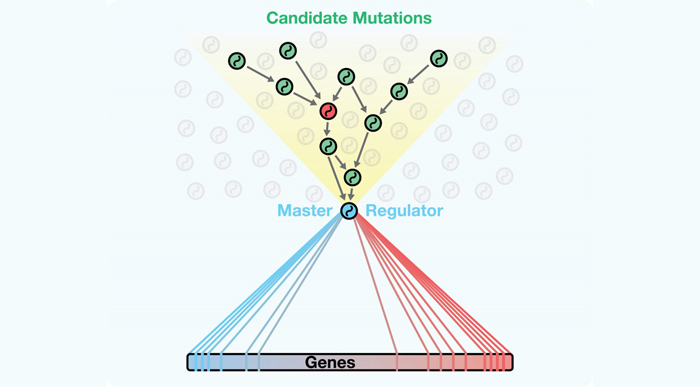
A new algorithm called DIGGIT identifies mutations that lie upstream of crucial bottlenecks within regulatory networks. These bottlenecks, called master regulators, integrate these mutations and become essential functional drivers of diseases such as cancer.
Although genome-wide association studies have made it possible to identify mutations that are linked to diseases such as cancer, determining which mutations actually drive disease and the mechanics of how they do so has been an ongoing challenge. In a paper just published in Cell, researchers in the lab of Andrea Califano describe a new computational approach that may help address this problem.
The manuscript presents an innovative algorithm called DIGGIT (short for Driver-Gene Inference by Genetic-Genomic Information Theory), which utilizes and adds an important new method to the toolbox the Califano Lab has been assembling over the past 10 years for modeling and interrogating regulatory networks. The fundamental idea behind it is that any mutation that represents a causal driver of a disease must be upstream of the “master regulator” genes that are the functional drivers the disease. (Master regulators are genes whose activity, either alone or in synergy with other genes, is essential for the onset and persistence of diseases such as cancer.) By thinking of master regulators as a “funnel” through which the pathways connecting the mutation to the disease phenotype are obligated to pass, DIGGIT can systematically identify the relatively small number of mutated genes that could drive disease.
Using this approach, the team discovered that loss of the gene KLHL9 drives the mesenchymal subtype of glioblastoma (GBM), the most deadly type of brain tumor. They also validated this finding through extensive experiments and in additional cohorts. Specifically, the team found that tumor growth in mesenchymal GBM lacking KLHL9 can be suppressed by reintroducing the gene. DIGGIT also accurately identified disease drivers in breast cancer and Alzheimer’s disease, suggesting that the algorithm, combined with MARINa, the Califano Lab’s extensively published tool for identifying master regulators of complex disease, now offers a powerful new method for identifying genetic drivers of other types of disease as well.
“We see this paper as a culmination of our efforts,” Dr. Califano says, “because it shows that in diseases as diverse as Alzheimer’s and different types of cancers, there are regulatory bottlenecks that integrate upstream signals, and channel them into the aberrant activity of master regulators. These, in turn, activate the pathological programs that are necessary for the emergence of disease.”
How DIGGIT works

James Chen, who recently completed his PhD in the Califano Lab, is the first author of the study.
The genesis of DIGGIT began with James Chen, a recent PhD graduate and member of the Califano Lab, as well as the first author of the paper. In his second year as a graduate student he met with Dr. Califano, who suggested that he write a proposal describing what he wanted to study and how it would fit alongside the other activities and resources available in his lab. “I proposed that we should be able to integrate genetics and genomics to identify driver mutations in cancer,” Dr. Chen recalls. “We hypothesized that the search for these driver mutations could be improved by integrating concepts from network biology and by building on the Califano Lab’s algorithms for identifying master regulators of disease.” Califano encouraged him to get started, and with the support of other researchers in the lab, they led the concept’s evolution into DIGGIT.
In the mesenchymal glioblastoma study, Chen began with gene expression and mutational profile data of more than 250 patients collected by the Cancer Genome Atlas consortium. The first step in the algorithm is to perform a genome-wide analysis to eliminate any genomic copy number variations (CNVs) that are clearly incapable of perturbing the molecular network of the tumor. DIGGIT filters the data to retain only CNVs at loci that are predicted to have cis-regulatory effects (changing gene expression of the genes in which they are located). This important first step eliminates CNVs that cannot cause changes in gene expression in the tumors, thus drastically amplifying the statistical power of any given collection of patient samples. In this way it overcomes a limitation of traditional genome-wide studies, which require large cohorts or large effect sizes to produce statistically robust results.
The remaining “functional” variants, or fCNVs, are then checked to see if they are predictive of increased activity of the master regulators connected with disease. DIGGIT evaluates the fCNVs using ssMARINA (single sample Master Regulator Inference Algorithm, another tool developed in the Califano Lab), which provides a quantitative measure of how active each master regulator is in each patient sample in the cohort. fCNVs that appear to alter master regulator activity are retained and then evaluated using MINDy (Modulator Inference by Network Dynamics), which predicts genes that are upstream of master regulators within the genetic network. Any fCNVs that are associated with a change in gene expression but are not connected to a master regulator in the interaction network are then eliminated from consideration. In this way, the combination of algorithms acts as a computational sieve, specifically capturing those genes that affect the activity of master regulators of a specific disease trait.
Andrea Califano explains how his lab's methods for identifying "bottleneck" genes in cellular networks can reveal biomarkers of disease. Video courtesy of Columbia University Medical Center.
The researchers hypothesized that the genetic origins of disease must lie upstream of master regulators that functionally drive the disease. The Califano Lab's study shows that looking at regulatory networks in this way can help to identify mutations that drive a cell's aberrant activity.
Finally, the best candidate driver mutations are identified using an analysis derived from classical genetic tests. The approach was developed to address the confounding issue that genomic alterations in cancer rarely occur individually. Rather, deletions and duplications take place across sections of chromosomes, leading to statistical dependencies between mutations; that is, genes that are next to each other are far more likely to be mutated together than genes that are more distant from each other. This also implies that any mutation that drives disease will be accompanied by additional mutations that are associated with disease only because of their proximity to the driver. To gain a clear picture of the genetic causes of disease, it is critical to distinguish the true driver mutations and artifacts of the analysis.
The team hypothesized that for any set of genes that are associated with a phenotype, no artifact gene can be more associated with that phenotype than the driver mutation. Using the list of candidate genes identified in the previous steps, the algorithm computationally assessed every gene in turn for its association with the mesenchymal GBM subtype. Through this process of elimination, DIGGIT narrowed the list of possible genes into a small list of loci that it predicted to be essential for the mesenchymal phenotype.
KLHL9 deletion drives mesenchymal glioblastoma
The DIGGIT pipeline identified two candidate genes that could be responsible for driving mesenchymal glioblastoma. The first, C/EBP-δ, had previously been identified in a collaboration with Antonio Iavarone, a professor of pathology and cell biology and neurology in the Columbia University Institute for Cancer Genetics, as playing a role in the development of mesenchymal tumors, so Chen focused on the second, which codes for a protein called KLHL9 that had never been identified as being relevant in brain cancer.
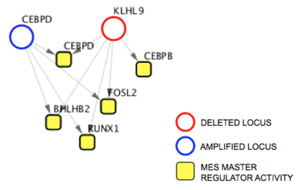
KLHL9 lies upstream of the master regulators C/EBP-β and C/EBP-δ.
A series of follow-up laboratory experiments demonstrated that loss of KLHL9 leads to aberrant activity in the mesenchymal master regulators C/EBP-β and C/EBP-δ. When normal KLHL9 function is restored, it suppresses the activity of these two genes by mediating the degradation of the proteins. Further tests explored the effects of KLHL9 in vivo by implanting mesenchymal glioblastoma cells into living mice. In mice in which KLHL9 expression was restored, tumor growth was significantly impaired.
Taken together with results of several additional experiments described in the paper, the findings reveal that deleting both copies of KLHL9 is sufficient to transform GBM tumors to an aggressive, mesenchymal subtype. Moreover, rescuing normal expression of KLHL9 is sufficient to severely hamper tumorigenesis of mesenchymal GBM. Considering that 50% of people with mesenchymal glioblastoma exhibit KLHL9 mutations, the findings suggest a potentially valuable therapeutic strategy for assessing brain cancer, by concentrating on the bottleneck that integrates the mutations as opposed to focusing on the mutations themselves.
Additional applications of DIGGIT
While implemented and validated mechanistically in GBM, DIGGIT can be used to investigate any phenotype for which matched gene expression and variant profiles (either somatic or germline) are available for a sufficient number of samples. To show that the algorithm could be used to identify genetic drivers of other diseases, Chen conducted additional analyses looking at BRCA-positive breast cancer and Alzheimer’s disease.
In the breast cancer study, he first searched the scientific literature for copy number variants already linked to breast cancer. He uncovered 25 alterations that had previously been reported as being associated with this breast cancer subtype. He then performed the DIGGIT analysis, comparing gene expression in breast cancer cells to that of normal breast tissue cells as controls. The algorithm identified 35 genes as drivers of BRCA breast cancer. Of the 25 genes previously identified in the literature, 19 (76%) appeared in the DIGGIT analysis, suggesting that the algorithm is highly capable of capturing driver mutations in other cancer types. It also revealed a number of never-before-seen genes that may warrant further investigation.
In the study of Alzheimer’s disease, DIGGIT identified 14 statistically significant variants that appear to drive the condition. Among these, the highest ranked was a variant in the gene TYROBP, which researchers at the Icahn School of Medicine at Mount Sinai independently predicted to be a driver of late-onset disease for the first time in 2013. DIGGIT also identified the APOE locus, a well-known variant associated with Alzheimer’s disease.
Because DIGGIT identifies mutations within the context of genome-wide regulatory networks, Dr. Chen points out that it offers an important advantage over traditional gene association methods. “Not only does looking at CNVs through the lens of master regulators dramatically increase our ability to detect candidates even in highly heterogeneous populations,” he explains, “it also provides a direct mechanistic perspective on exactly how these genes initiate the disease. There are many instances in which the identification of a candidate gene via genome-wide association studies precedes elucidation of its mechanism by years. Even in our studies of breast cancer and Alzheimer’s disease, where the goal was simply to show that DIGGIT could identify candidates that are missed by more traditional methods, it provided the additional benefit of immediately identifying the key molecular regulators and pathways that the mutations likely work through to produce a disease.”
Now a joint postdoctoral scientist in the laboratories of Angela Christiano and Andrea Califano, Dr. Chen continues work to incorporate the algorithm into Bioconductor, a widely used bioinformatics platform. In the meantime, the Califano Lab has begun to incorporate DIGGIT into its pipeline for integrating genetics and genomics across all tumors, including 20 in the Cancer Genome Atlas.
— Chris Williams
Related publication
Chen JC, Alvarez MJ, Talos F, Dhruv H, Rieckhof GE, Iyer A, Diefes KL, Aldape K, Berens M, Shen MM, Califano A. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell. 2014 Oct 9;159(2):402-14.