News
DAMAGES: A Method for Predicting Rare Disease Risk Genes
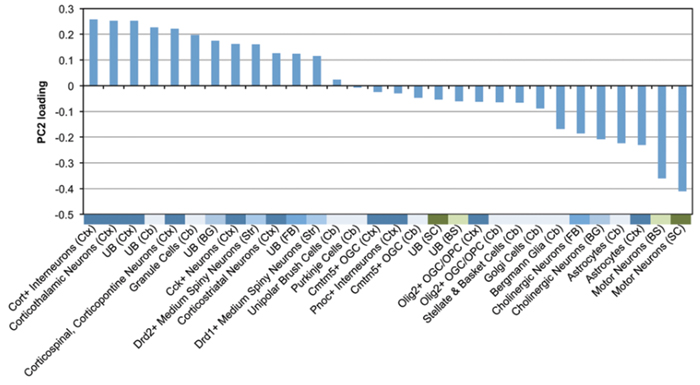
By inventing a new computational pipeline called DAMAGES, Chaolin Zhang and Yufeng Shen showed that brain cell types on the left of the plot are more prone to have rare autism risk mutations than cell types at the right. Narrowing the focus to these types of cells also helped to identify a molecular signature of the disorder that involves haploinsufficiency. Figure: Human Mutation.
Autism, a spectrum of neurodevelopmental disorders typically identified during early childhood, is widely thought to be the result of genetic alterations that change how the growing brain is wired. Nevertheless, despite a substantial effort in the field of autism genetics, the specific alterations that place one child at greater risk than another remain elusive. Although the list of alterations associated with autism is growing, it has been difficult to conclusively distinguish those that truly increase disease risk from those that are merely coincident with it. One troubling reason for this is that research so far seems to indicate that specific genetic abnormalities associated with autism risk are extremely rare, with many being found only in single patients. This has made it hard to reproduce findings conclusively.
In a paper recently published in the journal Human Mutation, Department of Systems Biology faculty members Chaolin Zhang and Yufeng Shen describe a method and some new findings that could help to more precisely identify rare autism-driving alterations. A new analytical pipeline they call DAMAGES (Disease Associated Mutation Analysis using Gene Expression Signatures) uses a unique approach to identifying autism risk genes, looking at differences in gene expression among different cell types in the brain in order to focus more specifically on mechanisms that are likely to be relevant for autism. Using this approach, they identified a pronounced molecular signature that is shared by disease risk genes due to haploinsufficiency, a type of genetic alteration that causes a dramatic drop in the expression of a particular protein.
The project described in the paper originated in Chaolin Zhang’s lab, which is interested in studying mutations that disrupt the function of RNA binding proteins, a class of molecules that regulate protein production in nerve cells. In the course of his research, Zhang noticed that some well-known autism associated genes exhibit different levels of expression in different cell types in the brain. Intuitively this makes sense, as our brain consists of many different types of nerve and supporting cells, and different kinds of cells perform different functions. Thus, one would expect them to operate differently at the molecular level. Zhang wondered whether this phenomenon could help in distinguishing pathogenic mutations from background mutations that are an ordinary byproduct of normal cellular replication.
In the case of autism, Zhang suspected that not all cell types are likely to be involved in producing the behavioral traits typical of the disease. Motor neurons, for example, are important in controlling muscle contractions and so play roles in ALS (also known as Lou Gehrig’s disease) and other neurological conditions that affect how the body moves. In a cognitive and behavioral disorder like autism, however, this type of function is less relevant. Instead, he hypothesized, one might expect that the changes in gene expression that would be most relevant would be found in cells that are involved in higher level cognitive functions.
To explore this hypothesis rigorously, Dr. Zhang worked with Yufeng Shen, a computational biologist who specializes in developing statistical methods for identifying rare genetic risk variants in limited data sets. The scientists realized that if the changes in gene expression that are important in autism take place only in specific cell types, the number of potentially relevant alterations becomes much smaller. Instead of looking for global changes in gene expression across the entire genome and in all cell types, it becomes possible to focus on a much more restricted data set, increasing statistical power to detect rare risk mutations.
“The more common approach that people are pursuing to identify rare mutations,” Zhang says, “is to develop large cohorts of autism patients and then use whole genome or exome sequencing to analyze their DNA. The problem is that gaining the necessary statistical resolution using that kind of an approach would require an enormous data set that would be extremely expensive to generate. By looking at gene expression — the RNAs that are being produced by cells — one by one in different cell types, we basically implement a filter that can quickly rule out mutations that are less likely to be relevant. This information allows us to constrain our search space dramatically, giving us higher specificity in our predictions, as well as a more refined picture of the mechanisms we should be paying more attention to.”
Based on this reasoning, Zhang and Shen used DAMAGES to score autism-associated changes in gene expression across each of 24 different cell types representing all major brain regions. For each cell type they compared samples representing autism with those from normal controls, making it possible to identify the changes in expression that were most associated with autistic phenotypes.
As they hypothesized, the researchers found that the genetic alterations with the highest DAMAGES scores seemed to concentrate in specific cell types. Although the method is not biased to look at one cell type differently from another, they discovered that brain cells that are most important in controlling functions such as attention, awareness, perception, and language — all features that become dysfunctional in people with autism — were most likely to exhibit changes in gene expression. This included cortical neurons, cerebellar granule cells, and striatal medium spiny neurons.
The scientists also examined the pathways in which the genes with the highest DAMAGES scores are thought to function. Such analysis connects expression signatures that are enriched in a particular sample to genetic functions in which those particular genes are thought to play a role. Zhang and Shen discovered that genes whose expression changed most dramatically are involved in processes such synapse formation, gene transcription, chromatin modification, and regulation of RNA metabolic process. These findings, they suggest, support other evidence indicating that autism is a disorder of transcriptional and post-transcriptional regulation of gene expression.
Moreover, when they looked in postmortem brain tissue from autism patients, the highest DAMAGES scores indicated a loss of gene expression, while increases in expression were virtually absent from the list of genes whose expression changed most significantly. The scientists postulate that this is the result of genetic alterations that cause haploinsufficiency. (Normally human cells have two copies of a gene, one from a person’s mother and one from the father. In haploinsufficiency one of these genes is severely damaged by “loss of function” variants or deletions, rendering the cell incapable of producing enough of a necessary protein.)

Department of Systems Biology Assistant Professsors Chaolin Zhang and Yufeng Shen collaborated on the development of DAMAGES.
In the paper, they report that this hypothesis is consistent with other recent findings by the Exome Aggregation Consortium (ExAC), which used an entirely different approach (large-scale sequencing of coding regions of DNA) to determine tolerance for rare loss of functional variants in general populations without developmental disorders.
“ExAC metrics and DAMAGES scores are based on orthogonal sources of information,” Shen says. “The former is about genetic variants in a large population and the latter is about cell-type specific gene expression. This makes these two approaches complementary and combining them would make it even more powerful.” In the end they focused on 117 mutated genes with the highest DAMAGES and ExAC scores, which indicated their high likelihood of increasing risk of autism, even though each gene has been found to be mutated in a single patient in the cohort they studied.
Although the paper does not completely solve the problem of definitively identifying rare autism risk genes, Zhang hopes that the approach it describes will encourage follow-up experimental research to begin testing its refined set of predictions. “As a complement to efforts that sequence entire genomes in large numbers of patients,” Zhang suggests, “we think that focusing on sequencing the loci we identified, and trying to find the same mutations in other patients, could be an efficient way to gain a clearer picture of the genetic risk factors and mechanisms that are truly at the root of the disorder.”
— Chris Williams
Related publication
Zhang C, Shen Y. A cell type-specific expression signature predicts haploinsufficient autism-susceptibility genes. Hum Mutat. 2016 Nov 16.