News
New Directions in Genome Engineering: An Interview with Harris Wang
As a graduate student in George Church’s lab at Harvard University, Harris Wang developed MAGE, a revolutionary tool for the field of synthetic biology that made it possible to introduce genomic mutations into E. coli cells in a highly specific and targeted way. Now an Assistant Professor in the Columbia University Department of Systems Biology, Dr. Wang recently published a paper in ACS Synthetic Biology that introduces an important advance in the MAGE technology. The new technique, called (MO)-MAGE, uses microarrays to engineer pools of oligonucleotides that, once amplified and integrated into a genome, can generate thousands or even millions of highly controlled mutations simultaneously. This new method offers a cost-effective way for designing and producing large numbers of genomic variants and provides an efficient platform for experimentally exploring genome-wide landscapes of mutations in bacteria and optimizing the organisms’ biochemical capabilities.
In the following interview, Dr. Wang explains the origins of the new technology, and discusses what he sees as the remarkable potential it holds for both basic biological research and industrial applications of synthetic biology.
How are MAGE and (MO)-MAGE different from more traditional methods in genome engineering?
In traditional genome engineering, researchers would induce genome perturbations randomly. For example, you might use ultraviolet radiation or a mutagen to generate mutations and then do a selection experiment to compare and isolate cells with different genotypes based on how they respond to specific stimuli. The problem with this approach, though, is that you have no way to control what mutations occur, even if you know the mutation you are interested in investigating.
In the late 1990s and early 2000s this led people to begin thinking about how to produce mutations in a targeted way. One important step forward occurred when Don Court and Barry Wanner independently developed a homologous recombination system using lambda red proteins. Their method enables double stranded recombination at a specific location much more efficiently than was possible beforehand. When I say “more efficient,” though, the efficiency was still only something like 10-4 (just 1 in 10,000 cells), and the technique only made it possible to induce a single mutation at a time.
Things started to move forward when Court discovered in the early 2000s that small oligonucleotides of 70-80 base pairs could be incorporated into the genome at very high efficiency by targeting them to specific sites of interest in homology arms. As a graduate student in George Church’s lab, I used this concept to design a way of doing targeted genome engineering across many different positions in the E. coli cell. You would design an oligonucleotide that, at either end, had a set of base pairs that was complementary to the genetic site of interest and also included the mutation you wanted to insert. If you knock out the genetic pathway through which cells usually repair replication errors, these oligos can then bind with their target sites during replication and are incorporated into the genome approximately 25% of the time. By repeating this process multiple times, you could then quickly increase the percentage of mutated cells in the population. We called this technology MAGE, which stands for multiplexed automated genomic engineering. (Read more about MAGE here.)
How did (MO)-MAGE grow out of this work?
After we developed MAGE, our next question was how to make it more cost-effective, which is critical because engineering a biological pathway can often require mutating a large number of genes. If you wanted to target thousands of sites simultaneously, you would need to order thousands of oligos, which is far beyond the financial resources of any lab.
We thought of a few potential ways to solve this problem, but the most tantalizing one was based on synthesis from microarrays. Although microarray synthesis has typically been limited to fewer than 60 base pairs, Agilent and other companies have recently developed the capability to synthesize up to 230 base pairs at high fidelity. This offers a great opportunity, though when you receive these oligos on a microarray, they come in just picoMolar concentrations. Because MAGE requires large numbers of oligos, (MO)-MAGE (which stands for microarray oligonucleotide-MAGE) borrows methods from gene synthesis to amplify the numbers of oligos a million-fold. Using common amplification primers, we design and synthesize a pool of reactions that contains sublibraries within the pool.
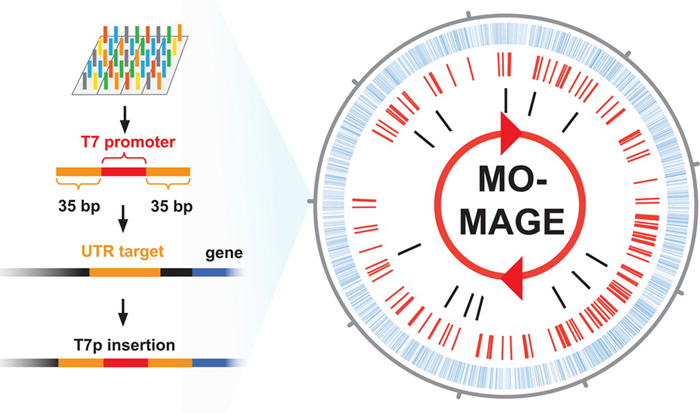
Wang and his collaborators demonstrated the feasibility of large-scale mutagenesis by inserting T7 promoters upstream of 2585 operons in E. coli using (MO)-MAGE. For each locus, the oligonucleotide promoter sequence was incorporated in a highly targeted way between two flanking regions that then incorporated it into single-stranded DNA. Using high-throughput sequencing they showed that all attempted insertions occurred at an average frequency of 0.02% per locus with 0.4 average insertions per cell.
In the (MO)-MAGE paper, for example, we describe a chip we designed that simultaneously introduces 13,000 mutations. Each of these features was grouped into one of 8 different classes, each of which had a different function. For example, one class included oligos designed to knock out the open reading frames in E. coli by introducing a stop codon and then frameshift mutation. Another class inserted a T7 polymerase promoter from the T7 phage, which allows you to do orthogonal regulation by expressing the T7 polymerase elsewhere in the genome. Another class changed ribosomal binding sites into the canonical sequence in order to tune up the translation initiation rates. Still another did the inverse by tuning down the translation initiation rates.
Using this pooled microarray-based approach, (MO)-MAGE is incredibly cost-effective and makes it possible to do large-scale genome engineering that would be impractical any other way. I calculated that if we had to make the equivalent number of reactions on a microarray chip using column-based synthesis, it would take something like $7 million and at least 5 months of work just to get the raw DNA. We can now do the equivalent work for a couple of thousand dollars in just a couple of weeks.
How do you know which oligos to insert in order to engineer the genome in specific ways?
If you want to target thousands of sites simultaneously, you’re not going to be able to select the oligos that target them by hand. And so as we were developing (MO)-MAGE, we also worked in collaboration with Morten Sommer at the Technical University of Denmark (DTU) to develop a computational design tool called MODEST. This web-based tool allows you to upload the positions and identities of a set of mutations across the genome that you want to make, and then uses an algorithm to generate a table of all of the oligo sequences you need to produce those mutations. You can then take that table to your favorite microarray synthesis company and after a couple of weeks they will send you back a tube containing those oligos.
How does being able to generate such large numbers of targeted mutations change the research that’s now possible?
The biggest problem with random mutagenesis is that the likelihood of a finding a beneficial mutation is astronomically low. (MO)-MAGE is not random, but it’s not a completely rational approach to engineering either. I like to think of it as a semi-rational approach whose beauty is that by allowing you to make many genetic variants very quickly, it opens up experimental opportunities that we’ve never really had before.
For example, computational analysis or the scientific literature might lead you to hypothesize that 5 genes are relevant in a specific biochemical process you are trying to optimize. But those genes exist within a complex molecular system and so identifying the ideal levels for all of these components in combination using traditional approaches poses a very difficult problem. By using (MO)-MAGE, however, you can quickly produce lots of genetic variants that you can just experimentally isolate and characterize. This allows you to tune the expression of all of the genes in an iterative way.
If you think about the traditional engineering pipeline that goes from design to building to testing, using this kind of semi-rational approach removes a historical bottleneck. Previously you might have been able to propose a variety of possible designs to optimize a specific biochemical activity, but it was never practical to build them all. (MO)-MAGE saves you from needing to put all your eggs in one basket with one design; it gives you a method to experimentally try hundreds of thousands or even millions of mutations and see what looks interesting. We’ve fixed that part of the pipeline.
In doing so, however, we’ve identified a new bottleneck, which is the question of how you systematically analyze the results of all of these mutations. We can build libraries of DNA very efficiently, but how do we study them in an equally efficient way? Are there genetic tricks or selection tricks that you could use to quickly pull out the most relevant mutations? These are the big questions the field is facing right now, and finding good answers would be very helpful for identifying or producing small molecules or materials of interest.
In what kinds of biological research is (MO)-MAGE going to be most useful?
What’s great about using this kind of semi-random approach is that it can often lead to new, unexpected turns in biology. If you’re only making one mutation at a time, you can only test one hypothesis at a time. But if you can generate 10 mutations, maybe 1 out of the 10 will give you an unexpected result. This offers a unique opportunity for further investigation.
Also, in addition to giving you information about the positive mutation space, large-scale perturbation using (MO)-MAGE also reveals the negative space. That is to say, some mutations might be really important to a particular biological problem, but because you can search the mutation space in a very comprehensive manner, you also eliminate everything that’s not important. For example, if you do a saturated mutagenesis of a region of 6 base pairs — in which you permute through every single base pair variation — you will have data on more than 4000 different variants, and you’ve completely covered that sequence. At that point, you should be able to say that you’ve experimentally validated your biological model or identify ways to make the model better.
(MO)-MAGE is also a particularly useful technique for understanding basic protein function because it allows you to target every single proton codon sequence one at a time. Traditionally, people have done this using alanine scans, a time consuming process in which you change every single amino acid to an alanine one at a time. If the change causes the protein to lose function you know that position is critical to the function of the protein. But using (MO)-MAGE we can now introduce pools of oligos such that within each pool you are not only scanning for alanine, but also for the other 19 amino acids. You can create a huge library of protein variants in which you assign mutations to each position, or multiple positions simultaneously, in a way that allows you to think about protein function holistically. To investigate the temperature stability of a protein, for example, you could generate a complete set of possible variants, subject them to heat stress, and then see which of those proteins remain stable.
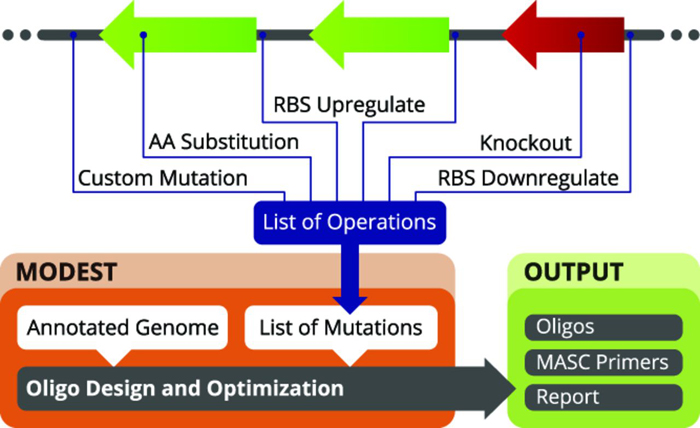
MODEST is a computational tool that can be used in conjunction with (MO)-MAGE. The user can select or supply an annotated genome and specify the desired mutations, which might include insertions, deletions, point mutations and amino acid substitutions, or phenotypic changes such as changes in the rate of protein translation and gene knockouts. MODEST processes this into a list of mutation objects, which are passed to an oligo design and optimization routine. This results in the design of MAGE oligos, MASC PCR primers, a report, and visualization of results.
In a project that’s in the pipeline right now we are using (MO)-MAGE to target essential genes. These genes are hard to manipulate with almost any other method; you can’t insert an antibiotic cassette in the middle of the essential gene because it will kill the cell. But by using these oligo pools you can essentially tile all the mutations you would be interested in making across the entire essential gene. And because each of the mutations could potentially affect the fitness of the cell, once you’ve made mutations across all of these positions you can then compete the variants together in a pool. The ones that are least disrupted will grow the fastest and the ones that are most disrupted will grow the slowest. You can then quantify, using deep sequencing, the relative frequencies of all of those variants across a pool. This would let you reconstruct a map of the mutation effect of this protein in one fell swoop.
Many scientists are interested in thinking about how you get from protein A to protein B. That is, what are the mutations that need to occur to make one protein into a substantially different protein? Often we identify proteins based on sequence homology, but sequence homologies for the most part are very large. Even when two proteins have homologous regions, they also have many regions that are very different. (MO)-MAGE could allow you to start thinking about how to approach this problem by iteratively making mutations and measuring the change in function in the resulting protein variants. As you walk through the mutation space in a semi-direct way, you can then start to map the genetic landscape of mutations that bridge the paths between two proteins of different function.
Industry has taken a lot of interest in synthetic biology. What do you see as some of (MO)-MAGE’s potential commercial applications?
I recently presented (MO)-MAGE at a conference and people were very excited about the prospects of doing large-scale genomic perturbations at the industrial level. It took DuPont 12 years and something like $300 million to engineer a strain of 1,3-Propanediol that’s used today as an additive in many applications, from carpeting to paint thinners. They make millions of tons of this stuff in huge fermentation reactions, and to optimize the process they introduced something like 37 mutations. In contrast, we can now generate hundreds of thousands of targeted mutations for a small fraction of this time and cost.
If you’re an engineer working in industry, once you’ve identified a biochemical pathway that produces a desired product, the next challenge is to optimize it. Traditionally, this process has been very clunky, requiring a lot of labor-intensive trial and error. Now you can just make a list of the genes and mutations that might be important and try all of the possible combinations. Each gene in one of these multi-gene pathways is like a knob, and (MO)-MAGE allows you to dial the strength of those individual knobs to identify the configuration of genetic mutations that generates the maximum flow of resources through that pathway.
Scientists and engineers who work in industry are also concerned about things like pH resistance, solvent resistance, temperature resistance, and other things that constitute global changes whose origins are unclear. Some mutations that are seen in the development of these kinds of resistance are hitchhiker mutations, while others are key driver mutations. Using (MO)-MAGE you could conceivably look at parallel strains with independent mutations and then generate a hybrid version of those two strains in order to incorporate combinations of mutations. Evolution working on its own may not have had time to access those mutations in concert, but as synthetic biologists we can. It gives us the opportunity to leapfrog to other beneficial genetic states.
In industry they say that time is money, so whatever you can do to reduce the cycle time of discovery and development, or to increase the efficiency through which a strain of bacteria generates your product, will make a big difference, particularly if you’re scaling your production up to millions of liters.
What’s next for (MO)-MAGE?
We are definitely interested in further developing (MO)-MAGE so it could be applied to other scientifically relevant organisms. We designed MAGE to engineer E. coli, but it could also potentially be used to look at the molecular origins of virulence in pathogenic E. coli and things of that sort.
My lab is also thinking about how these approaches could be used to engineer the gut microbiome. For example, we’re thinking about strategies for generating oligos inside the cell instead of having to deliver them. If you could have the cells produce them naturally you could potentially improve the incorporation efficiency. These kinds of advances would add to the (MO)-MAGE repertoire from a technology perspective.
There’s also lots of really interesting biology that could come out of this. For example, scientists have identified many mutations through experimental laboratory evolution. But the question is, are all of those mutations important? What are those mutations doing? Do those mutations have to work in concert or do they work individually? Are mutations additive or do they have synergistic or antagonistic effects? Obviously you can’t “rewind” evolution, but in a certain way (MO)-MAGE offers that opportunity. We can now introduce specific mutations into an ancestral strain in any combination we want, in any order we want, creating a very directed evolution. Essentially, this gives you a way to do retro-evolution through forward engineering, letting you unwind the evolutionary process. It’s an application that is ripe for this type of technology.
— Interview by Chris Williams
Related publications
Bonde MT, Kosuri S, Genee HJ, Sarup-Lytzen K, Church GM, Sommer MO, Wang HH. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth Biol. 2014 Jun 20. [Epub ahead of print]
Bonde MT, Klausen MS, Anderson MV, Wallin AI, Wang HH, Sommer MO. MODEST: a web-based design tool for oligonucleotide-mediated genome engineering and recombineering. Nucleic Acids Res. 2014 Jul;42:W408-15.