News
Novel Method Identifies New Risk Genes for Developmental Disorders
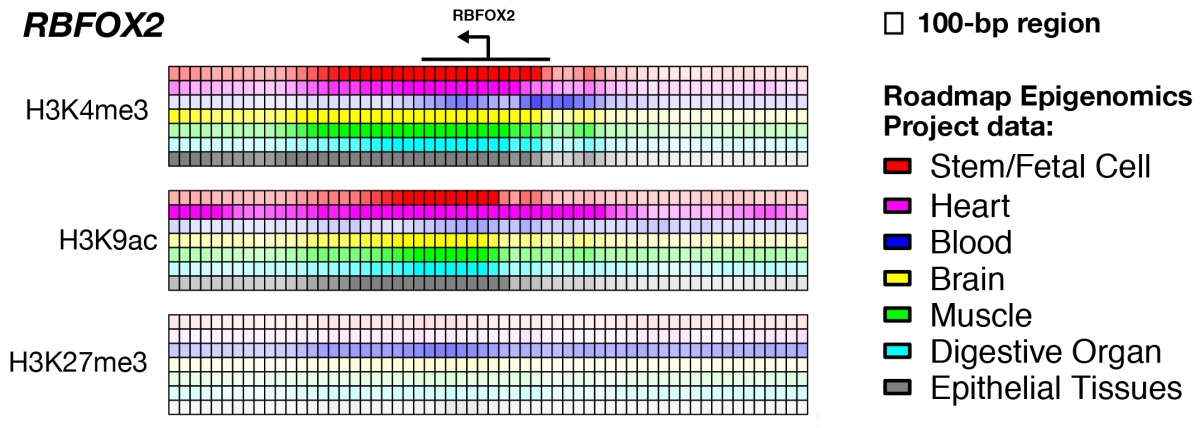
The epigenomic profile of RBFOX2, a haploinsufficient gene recently identified as a risk gene of congenital heart disease. Each small box represents 100 bp region around transcription start sites (TSSs) and the shade of the color reflect the strength of the histone mark signal in tissues under normal conditions. RBFOX2 has large expansion of active histone marks (H3K4me3 and H3K9ac), especially in heart and epithelial tissues (purple and gray rows), and tissue-specific suppression mark (H3K27me3) in blood samples.(Credit: Shen lab)
The genetics of developmental disorders, such as congenital heart disease and autism, are highly complex. There are roughly 500 to 1,000 risk genes that can lead to each of these diseases, and to date, only about a few dozen have been identified. Scientists have ramped up efforts to develop computational approaches to address challenges in accurately identifying genetic risk factors in ongoing genetic studies, and the availability of such tools would greatly assist researchers in gaining a deeper understanding of the root causes of these diseases.
Focusing on haploinsufficiency, a key biological mechanism of genetic risk in developmental disorders, Yufeng Shen , PhD, and his lab have developed a novel computational method that enables researchers to find new risk genes in these diseases. Their key idea is that the expression of haploinsufficient genes must be precisely regulated during normal development, and such regulation can be manifested in distinct patterns of genomic regulatory elements. Using data from the NIH Roadmap Epigenomics Project, they showed there is a strong correlation of certain histone marks and known haploinsufficient genes. Then based on supervised machine learning algorithms, they developed a new method, which they call Episcore , to predict haploinsufficiency from epigenomic data representing a broad range of tissue and cell types. Finally, they demonstrate the utility of Episcore in identification of novel risk variants in studies of congenital heart disease and intellectual disability.
Haploinsufficiency means that a loss of function of one of the two copies of a particular gene could cause human diseases. Previous studies have shown that developmental disorders are often caused by loss of function mutations in such haploinsufficient genes.
However, the critical challenge the researchers set out to address was to devise a way to computationally identify these haploinsufficiencies.
“We know that a lot of genes can be haploinsufficient, but we don’t know which of these genes are, in any given study,” says Dr. Shen, assistant professor of systems biology at Columbia University Irving Medical Center (CUIMC). “So, the question we sought to answer is whether we can pre-compute or predict which genes are haploinsufficient before we do an actual human genetic study.”
The researchers’ new method successfully pre-computes whether or not a gene is likely to be haploinsufficient; Once this information is determined, researchers can zero in on the genes in a study that are likely to be haploinsufficient and further analyze the loss of function variance only in that very precise dataset.
“If a gene is determined haploinsufficient and we are able to observe loss of function in the patient, then it is likely that this is a risk gene,” explains Dr. Shen. “Episcore enables us to increase our chances of being able to predict which haploinsufficient gene could be a risk gene. Furthermore, we can combine this with additional statistical analyses to implicate new risk genes.”
The researchers compared Episcore’s performance to that of existing methods. Based on data from recent exome sequencing studies of developmental disorders, Episcore achieved better performance overall in prioritizing loss of function de novo variants, or new mutations, than current methods. One of the reasons for the better performance is that Episcore, notes the researchers, is not biased towards well-studied genes, because the data used for the method were generated by throughput technologies without preference to well-studied genes. Some previous methods used protein-protein interaction networks or gene pathways as input, which are inevitably biased toward well-studied genes. One of the most popular competing methods, ExAC pLI, is based on depletion of rare genetic variation in general population. The epigenomic data used by Episcore are orthogonal to population genetic data, making Episcore and ExAC pLI to be complementary with each other.
Episcore is detailed in the paper , “Distinct Epigenomic Patterns are Associated with Haploinsufficiency and Predict Risk Genes of Developmental Disorders”, published in Nature Communications. Columbia University coauthors with Dr. Shen, include Xinwei Han (former postdoc research scientist in the Shen lab), Siying Chen (systems biology PhD student in the Shen lab), Elise Flynn (systems biology PhD student who rotated in the Shen lab), Shuang Wu (biostatistics master student), and Dana Wintner (summer undergraduate student from Cornell University).
The work was funded by NIH grant R01GM120609.
-Melanie A. Farmer