News
Systems Biology Throws You Out of the Box: An Interview with Saeed Tavazoie

One of the defining features of systems biology has been its integration of computational and experimental methods for probing networks of molecular interactions. The research of Saeed Tavazoie, a professor in the Columbia University Department of Systems Biology, has been emblematic of this approach. After undergraduate studies in physics, he became fascinated by the processes that govern gene expression, particularly in understanding how gene expression is regulated by information encoded in the genome. Since then, his multidisciplinary approach to research has generated important insights into the principles that orchestrate genome regulation, as well as a number of novel algorithms and technologies for exploring this complex landscape.
In this conversation, Dr. Tavazoie discusses his research in the areas of gene transcription, post-transcriptional regulation, and molecular evolution, as well as some innovative technologies and experimental methods his lab has developed.
How would you describe your approach to biological research?
Traditionally in biology, you had to take a very focused view of a problem. You might look closely at a particular protein or a specific biological process, and after reading a lot of papers about it you could begin asking a question and get an answer that moved our understanding of that problem a little bit forwards. In systems biology, however, we are not interested in one particular protein or process, but in identifying the underlying principles of how the entire system behaves. This perspective has been made possible by a number of new technologies that now allow us to make large-scale measurements on the entire system — including not just all of the genes and proteins, but all of the interactions between them. Once we gather these large collections of data, we can then analyze them to reverse engineer how all of the components within a regulatory network come together to orchestrate cellular behavior.
"It’s not just that you start thinking out of the box, new technologies actually throw you out of the box and you can’t avoid thinking about things in new ways."
This approach brings about an important change in perspective. Instead of imposing a set of expectations that define how we think biology should behave, we try to be as unbiased as possible and let the system tell us what’s interesting. This isn’t to say that the traditional methods aren’t important, but when an entire system behaves in ways you never anticipated, that’s when important new discoveries are made. Although it can be useful to explain how one molecule interacts with one other molecule, the goal should be to walk away from your experiment learning a general principle that goes well beyond this specific interaction. In almost every experiment we do in our lab we try to set things up so that it could potentially produce these kinds of insights.
The new technologies that we and others are developing not only generate a scaffold of knowledge about regulatory interactions that other scientists can use, but eventually become important tools for making progress in many other areas. In the past, technologies like microarrays, RNA-Seq, and CHiP-Seq totally changed the way people do science. Today, new technologies that are coming out of systems biology are pushing conceptual revolutions in biology because they enable you to make observations you couldn’t make before. It’s not just that you start thinking out of the box, new technologies actually throw you out of the box and you can’t avoid thinking about things in new ways.
Can you give an example of a technology that you are working on and how it is changing your perspective on biology?
One area of research that has seen a lot of activity in recent years is to study how transcription factors regulate gene expression. Transcription factors are proteins that bind to DNA and either promote or repress DNA’s transcription into RNA. There is a technology called CHiP-Seq that uses antibodies that can recognize and bind to specific transcription factors. You can fragment the DNA into millions of pieces and then use an antibody to find all the places in the genome that are bound by this protein, giving you a snapshot of whether or not a particular gene is being regulated at a particular time point.
Although this is a powerful approach, it can only tell you the binding locations of a single kind of transcription factor at a time. Some years back I wondered if it might be possible to develop a technology that would let us identify the binding sites for all of the hundreds of proteins that are bound to different segments of the genome at a given time, all at once. In our lab we have now developed a technology that can do this. Using a series of purifications of protein-DNA complexes, we can separate naked DNA from DNA that has proteins bound to it, process these segments, and then use high-throughput sequencing to identify the chromosomal location for each of the bound transcription factors, giving a global profile of protein occupancy throughput the genome.
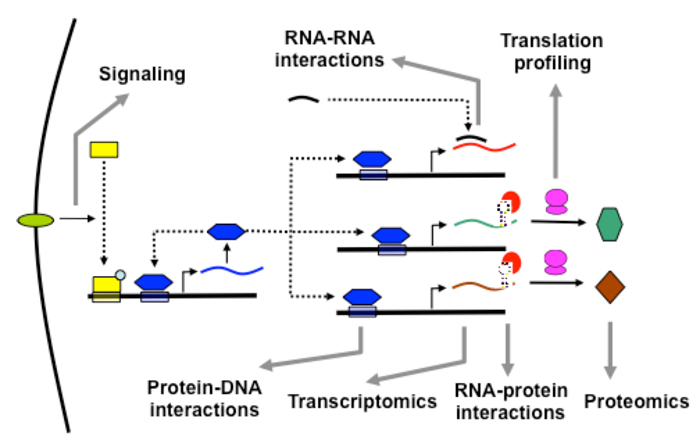
The Tavazoie lab studies multiple layers of molecular interactions in an effort to develop systems-level predictive models of cellular behavior.
This technology, co-developed with a former graduate student, Tiffany Vora, allows us to identify all the sites in the genome that are bound by protein, but by itself can’t tell you the identity of each protein. To solve this problem, Peter Freddolino, a postdoctoral researcher in my lab, developed a computational method that takes this readout of protein occupancy and, using a modification of an algorithm called FIRE that my lab developed previously, generates binding sequence models of where the proteins are found. So now you can go in and systematically learn from these experiments what the sequence preferences are for a large number of proteins at once.
Having this technology opens up a number of exciting possibilities, particularly when it is combined with RNA-Seq, which gives a complete picture of the RNAs in the cell at a given time. If you perturb the cell in some way it now becomes possible to monitor all of the locations in DNA that are bound by transcription factors, while simultaneously recording the entire profile of RNAs that are present (also called the transcriptome). Because the networks of interactions among these different components are dynamic, we take a series of measurements every few minutes, monitoring how these interactions evolve over time. We want to get to the point where you could basically make a movie consisting of a series of snapshots of which proteins are binding to which genes and when the RNAs are being made. It would be really enabling for systems biology, giving us the kind of observations that we really want. And most importantly, it would be mechanistically anchored in physical interactions.
What would be some of the potential applications of this kind of approach?
In our initial tests, we focused on E. coli, a bacterium that has been studied for about 70 years and whose binding specificities are largely known. When we use our approach without using any of these earlier findings as input, we rediscover those binding sites that people discovered over decades. But whereas it required years of intensive laboratory work to find these binding events in the past, we identify them, and others, in just a few experiments.
Now that we know this works in E. coli, we can begin learning binding site preferences in other organisms we don’t know anything about. The vast majority of bacteria that cause infectious diseases and play important roles in ecosystems have not been studied at the level of detail we now have for E. coli, so we know very little about their regulatory networks. We can now take bacteria that are important but not well studied, run them through our pipeline, systematically annotate all of their binding sites, and generate binding site models that would rapidly expand our knowledge about their regulatory networks.
That kind of result would be significant in and of itself, but our approach also produces knowledge about regulatory interactions inside cells that will be useful to scientists who work in pathogenic systems, no matter what they’re studying. These regulatory networks are engaged in almost any process someone might look at, and react to any kind of perturbation you might study, like oxidative stress or antibiotic stress. Knowing the mechanisms through which regulatory networks function is a huge step toward figuring out what’s going on. This will be really powerful.
How about other layers of regulation, such as post-transcriptional regulation? How do they fit into the picture?
Although a big focus for systems biology in its first 10-15 years has been on transcription, we and others are discovering that there’s also a huge amount of regulation that occurs after the messenger RNA (mRNA) has been created. In the regions at the ends of the transcript called 5’ and 3’ untranslated regions, for example, the mRNA can be bound by proteins that can increase the degradation rate of the mRNA. Discovering these elements in RNA is more challenging than in DNA because RNA is single stranded and can form secondary structures where the proteins bind. All of this means that even if studies along the lines of what I was talking about earlier determine that you are highly expressing a gene at a particular moment, the transcript could be degraded very quickly and might not actually play the role that those findings might indicate.
"Much of post-transcriptional regulation is still a black box. We're trying to change that."
Over the last few years we have developed computational methods that look through the entire transcriptome and discover regulatory elements in RNA. Basically, we simulate RNA binding by testing hundreds of billions of possible structures in the computer and calculate which ones have a high likelihood of being involved in post-transcriptional regulation. We need massive computational resources to sift through all of them and identify ones that are functionally important. To do one run on a data set can take 3-4 days using 400 processors on the Department of Systems Biology’s Titan cluster. That’s a huge amount of computing that would be hard to do without having this kind of infrastructure here at Columbia.
We’re finding that post-transcriptional regulation plays important roles almost everywhere we look. The challenge now is not only to catalog the protein binding sites, but also to find the proteins that recognize them, figure out what the proteins are doing, and see what other regulators a protein is interacting with; this knowledge is necessary to work out the entire pathway of regulation.
It’s exciting because if you think about cancer biology, for example, over the years people have discovered that a large number of oncogenes and tumor suppressors are transcription factors. But we’re discovering that gene expression is modulated not only by how much RNA you make, but also by how fast the mRNAs degrade, because if you degrade an mRNA it doesn’t have a chance to generate a protein. There are actually two regulatory inputs, and understanding how they interact is going to provide a clearer picture of what’s happening at the molecular level. Much of post-transcriptional regulation is still a black box, though, because people have not been able to study it effectively so far. We're trying to change that.
Do these regulatory networks tend to be stable, or do they change over time?
Our lab is actually very interested in the principles that govern how regulatory networks change over very long evolutionary time scales. We study this by carrying out experimental evolution in the laboratory. The nice thing about working with bacteria is that they divide once every hour, so over the course of weeks to months, you see a large number of generations. In our experiments we can expose them to different extreme environments and see how cells survive and adapt to the challenges that the new environments create, look at what kinds of modifications occurred in the genome, and determine how they affected the regulatory network.
One of the things we’ve discovered is that it seems to be much easier than we had previously thought for bacteria to adapt to extreme environments, and they seem to do so by rewiring their regulatory network. Historically, most people have thought that adaptation to new environments happens by making changes in the coding regions of the genome and that these changes generate a protein that functions better in the new environment, improving the organism’s fitness. People have thought that these kinds of positive improvements in fitness and adaptation happen through very gradual, subtle, rare changes in amino acids at the protein level and take a long time to happen.
What we’re discovering, though, is that upon the transition to these extreme environments, the dominant mutation mechanisms are not the gain of new functions through advantageous mutations, but rather loss-of-function mutations in genes that play regulatory roles. That is, the organisms adapt when one gene is lost that lets another gene that it had previously suppressed become activated. That’s the nature of regulatory networks.
Although it wasn’t initially obvious, this makes sense because it’s very easy to mutate a gene in ways that will make a protein stop functioning. It’s much, much harder to get subtle, rare mutations that actually enable the bacteria to survive better. This is because the rich regulatory network that exerts control over all the genes evolved in the native habitat of the organism. When you take the organism outside that context, there’s no reason to expect that the regulatory network is going to do the right thing. It’s like taking an organism with a cognitive system and putting it in a completely weird psychological environment. It’s going to go crazy. It’s not going to adapt. That’s the way we think about it now. And we’re seeing a huge amount of this, which is very exciting.
We’re exploring how this perspective could help to explain antibiotic resistance. Within the clinical setting, there has been great concern because, increasingly, bacteria can survive treatment with antibiotics that were once able to eliminate infections. We would like to identify the major antibiotic mechanisms by discovering how bacteria adapt to environmental challenges and evolve novel pathways of resistance.
You compared this exposure to extreme environments as producing a cognitive response. Is that to say that bacteria can think?
Obviously they can’t think in the same way that humans think, but a few years back we applied a systems biology approach to looking at bacterial behavior. We discovered that bacteria make predictions about their environment, very much like nervous systems do. They actually anticipate what’s going to happen next.
When we raised the temperature at a constant concentration of oxygen on a culture of E. coli, we saw that all the genes involved in aerobic metabolism went down in a synchronized way. We initially found this nonsensical because it isn’t advantageous to shut down aerobic metabolism in the presence of oxygen. But the changes make sense in the context of the bacteria’s native habitat if you look at the ecology of the organism and what it’s been experiencing over geological time scales. When E. coli enter the mouth, the temperature rises to 37°C. Then they go down the gastrointestinal tract and after 10-20 minutes oxygen levels drop. So when the temperature goes up, the networks start working in anticipation of the change in oxygen levels. This kind of anticipatory behavior was not known in cellular systems before.
Being minimally biased puts you in a domain of exploration that you couldn’t have anticipated ahead of time.
The only way we could see this was by looking at the entire gene expression dataset, and so this is a great example of the power of the systems biology approach. Because it’s minimally biased it puts you in a domain of exploration that you couldn’t have anticipated ahead of time. It’s not unlike the bacteria, actually, in that scientifically you get dropped into an environment you haven’t seen before and need to adapt your perspective to what the data are telling you. This approach has changed how we do biology in ways that are very exciting to be a part of.
— Interview by Chris Williams