News
Cancer Oncotecture: A Universal Model of Tumor Survival
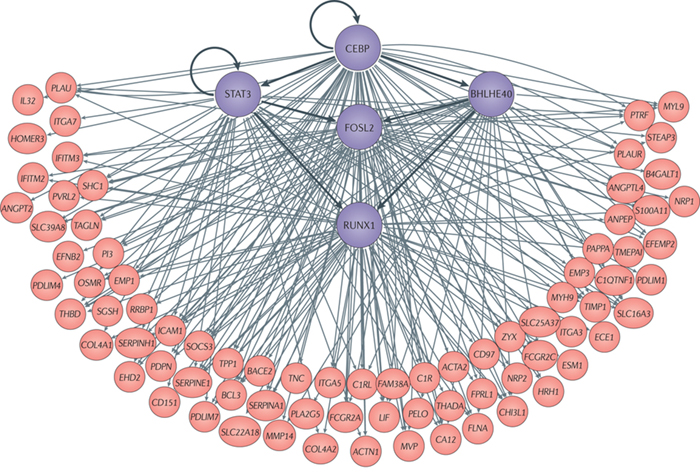
An example of tumor oncotecture. Transcription factors involved in the activation of mesenchymal glioblastoma subtype are shown in purple. Together, they comprise a tightly knit tumor checkpoint, controlling 74% of the genes in the mesenchymal signature of high-grade glioma. CEBP (both β and δ subunits) and STAT3 regulate the other three transcription factors in the tumour checkpoint, synergistically regulating the state of mesenchymal GBM cells. (Image: Nature Reviews Cancer)
In a detailed Perspective article published in Nature Reviews Cancer, Department of Systems Biology chair Andrea Califano and research scientist Mariano Alvarez (DarwinHealth) summarize more than a decade of work to propose the existence of a universal, tumor independent “oncotecture” that consistently defines cancer at the molecular level. Their findings, they argue, indicate that identifying and targeting highly conserved, essential proteins called master regulators — instead of the widely diverse genetic and epigenetic alterations that initiate cancer and have been the focus of much cancer research — could offer an effective way to classify and treat disease.
As coverage of the paper in The Economist reports:
ONE of the most important medical insights of recent decades is that cancers are triggered by genetic mutations. Cashing that insight in clinically, to improve treatments, has, however, been hard. A recent study of 2,600 patients at the M.D. Anderson Cancer Centre in Houston, Texas, showed that genetic analysis permitted only 6.4% of those suffering to be paired with a drug aimed specifically at the mutation deemed responsible. The reason is that there are only a few common cancer-triggering mutations, and drugs to deal with them. Other triggering mutations are numerous, but rare—so rare that no treatment is known nor, given the economics of drug discovery, is one likely to be sought.
Facts such as these have led many cancer biologists to question how useful the gene-led approach to understanding and treating cancer actually is. And some have gone further than mere questioning. One such is Andrea Califano of Columbia University, in New York. He observes that, regardless of the triggering mutation, the pattern of gene expression—and associated protein activity—that sustains a tumour is, for a given type of cancer, almost identical from patient to patient. That insight provides the starting-point for a different approach to looking for targets for drug development. In principle, it should be simpler to interfere with the small number of proteins that direct a cancer cell’s behaviour than with the myriad ways in which that cancer can be triggered in the first place. (Read full article.)
The Nature Reviews Cancer paper provides an overview of the Califano Lab’s efforts, summarizing its methods for master regulator analysis, along with evidence it has amassed in support of its model. The authors suggest that specific master regulator proteins and the tumor checkpoints they comprise function consistently as bottlenecks in the molecular networks that give rise to specific cellular phenotypes. In this way, they canalize diverse upstream genetic and epigenetic alterations to implement the gene transcriptional programs that activate cancerous activity, maintain cancer cells in a malignant state, and enable the development of tumor drug resistance. These proteins, therefore, constitute an “Achilles heel” of cancer, and offer an attractive, distinct category of biomarkers and therapeutic targets that complements other approaches.
Califano and Alvarez are careful to distinguish their strict concept of master regulators from an increasingly common and looser definition of the term among cancer biologists. Whereas some have used the term to describe genetic determinants or functional drivers of cancer, the authors emphasize that master regulator proteins are necessary and sufficient to produce a transcriptional signature that is distinctive to a particular cancer subtype and to regulate it directly. Adhering to this stricter definition, the paper argues, will provide better targets for selectively killing tumor cells.
The paper describes the opportunities made possible by the Califano Lab’s various computational algorithms, including ARACNe, MARINa, VIPER, and DIGGIT, which together comprise a framework for inferring molecular networks based on gene expression data, identifying tumor checkpoints in those networks, and developing a mechanistic understanding of tumor biology. It also explains the lab’s use of cross-species approaches to validate its predictions, as well as methods for connecting master regulators to drugs and drug combinations that are likely to be capable of targeting them.
In addition, the paper acknowledges some limitations and potential improvements to the paradigm. These include the need for better methods for distinguishing differences in regulatory networks between tissues and among single cells within the same sample, as well as for modeling kinetics of mRNA expression and protein abundance over time, for example following a drug perturbation.
Ultimately, the publication suggests that using cancer oncotecture as a framework for directly studying the activity of cancer proteins has many advantages. “Genetics approaches that pursue the oncogene addiction paradigm are quickly running out of low-hanging fruits,” Califano says. “We think that focusing on the protein level, and particularly what appears to be a relatively small number of master regulator proteins, could simplify the problem of identifying essential tumor dependencies and help identify better strategies for precision medicine.”
— Chris Williams
Related publication
Califano A, Alvarez M. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer. 2016 Dec 15.