News
New Insights on How the Reprogramming Factor LIN28 Regulates its Targets
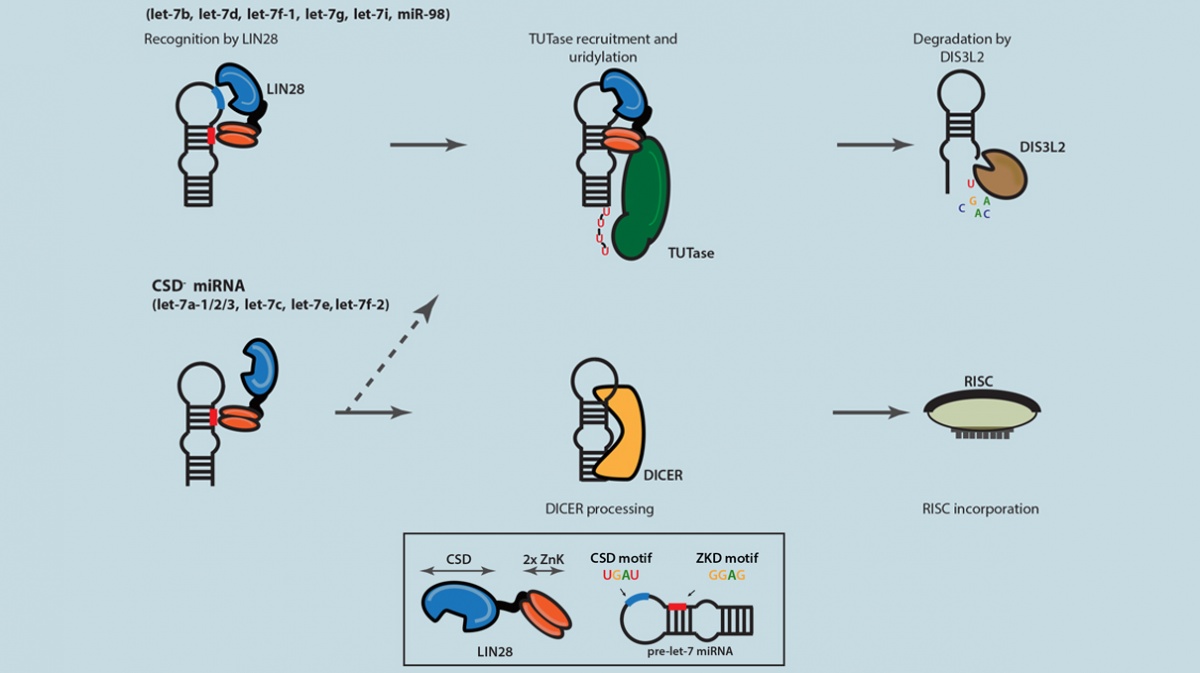
The proposed model of selective Let-7 microRNA suppression modulated by the bipartite LIN28 binding.(Image courtesy of Zhang Lab)
A new study led by Chaolin Zhang, PhD , assistant professor of systems biology , published today as the cover story of Molecular Cell , sheds light on a critical RNA-binding protein that is widely researched for its role in stem cell biology and its ties to cancer progression in multiple tissues.
The LIN28 RNA-binding protein, initially found in worms about 15 years ago, is specifically expressed in stem cells. It became well known because the protein is one of the four factors that were used to “reprogram” skin cells to induced pluripotent stem cells, or iPSCs, a breakthrough that was awarded the Nobel Prize in 2012. More recently, it was determined that the LIN28 RNA-binding protein can also be reactivated in cancer to drive tumor growth and progression. Due to its critical importance in developmental and cancer biology, scientists want to understand the role LIN28 plays at the molecular level. This new study provides some understanding of how the LIN28 protein suppresses a specific family of microRNAs, called Let-7, which are selectively lost in cancer.
“Let-7 microRNAs are the major downstream targets controlled by LIN28 identified so far. While LIN28 is mostly found in stem cells, Let-7 is only detected in differentiated cells because of stem cell-specific suppression by LIN28. However, the interplay between the two is still not well understood,” says Dr. Zhang, who is also a member of Columbia University’s Center for Motor Neuron Biology and Disease . “This study contributes to our understanding of how LIN28 suppresses Let-7, as well as provides a refined model for this important, rather complex molecular pathway.”
MicroRNAs, also referred to as miRNAs, are a class of small regulatory RNAs that are involved in essentially all cellular processes. Let-7 miRNA family, the focus of this particular work, is an ancient family of miRNAs whose expression is required for proper developmental timing and tumor suppressor function.
Researchers have been focusing on understanding Let-7’s various functions due to evidence that links the loss of Let-7 to the development of aggressive cancers. It has also been uncovered that Let-7 has to be suppressed (by LIN28) for the self-renewal of stem cells. Interestingly, in humans and other mammalian species, there are 12 Let-7 family members that were generated by genomic duplications during evolution and fixed ever since. These members are thought to have the same functions and are all suppressed by LIN28 through the same mechanism. Still, the reason for 12 different copies remains a mystery.

Cover artwork for Molecular Cell, Vol. 71, Issue 2; Let-7 microRNA partially escapes recognition and suppression by LIN28. Art by Dmytro Ustianenko with inspiration from the mural, Balloon Girl (2002) by Banksy.
In the new study, Zhang and collaborators analyzed specific binding sites of LIN28 using their own computational method that maps protein-RNA interactions at the single-nucleotide level, mapping tens of thousands of LIN28 binding sites in mRNA derived from CLIP data (CLIP is a biochemical assay used in the field that enables the analysis of protein interactions with RNA on a genome-wide scale).
Their analyses revealed an entirely new RNA sequence pattern (aka motif) recognized by LIN28 in addition to another sequence motif that was previously known to bind LIN28. Careful characterization demonstrated that the new motif was recognized through a protein domain in LIN28 called cold-shock domain (CSD), while the other known motif was recognized through the zinc knuckle domain (ZKD) of LIN28. Excitingly, when the study’s authors re-examined LIN28 binding sites in the precursor forms of Let-7 miRNAs, they found that only half of Let-7 family members have CSD binding sites, and the other half do not, and because of this, LIN28 binds to the latter group much more weakly.
“When we examined the protein-RNA interaction data we found a striking difference in how robustly LIN28 will bind to Let-7 miRNA depending on whether they have this new sequence motif. This leads us to believe—and validate—that not all members of the Let-7 family are equally suppressed by LIN28,” says Dr. Dmytro Ustianenko, a postdoctoral scientist in the Zhang lab and lead author of the study. “I have been working on this pathway since the start of my PhD, and I am very excited to add another small piece to the solution of this complex puzzle.”
The researchers say their finding could lead to a new set of questions. “The selective suppression of Let-7 through this molecular switch provides a fine tuning mechanism. We found that some Let-7 fami ly members partially escaped from suppression in stem cells and cancer. This implies a potential new role of LIN28 and Let-7 in pluripotent and cancer stem cells,” notes Zhang and Ustianenko. The exact nature of such function still needs to be investigated in a future study, which could lead to better understanding why some cancers are more aggressive than others.
The study titled, “LIN28 Selectively Modulates a Subclass of Let-7 microRNAs,” was published July 19 in Molecular Cell. Dr. Zhang’s collaborators include coauthors Dmytro Ustianenko (Columbia University); Hua-Sheng Chiu (Baylor College of Medicine); Thomas Treiber (University of Regensburg, Germany); Sebastien Weyn-Vanhentenryck (Columbia University); Nora Treiber (University of Regensburg, Germany); Gunter Meister (University of Regensburg, Germany); and Pavel Sumazin (Baylor College of Medicine).
The study was funded by the National Institute of Neurological Disorders and Stroke and the National Institute of General Medical Sciences.
Primary research conducted in the Zhang lab focuses on development of the nervous system and its underlying molecular mechanisms. The group concentrates on the function of post-transcriptional gene regulation by RNA-binding proteins. Their work is based on mouse models and in vitro neural differentiation of embryonic stem cells, as well as a combination of high-throughput data-driven and hypothesis-driven approaches.
-Melanie A. Farmer