News
Novel Machine Learning Method Expands the Landscape of Breast Cancer Driver Genes
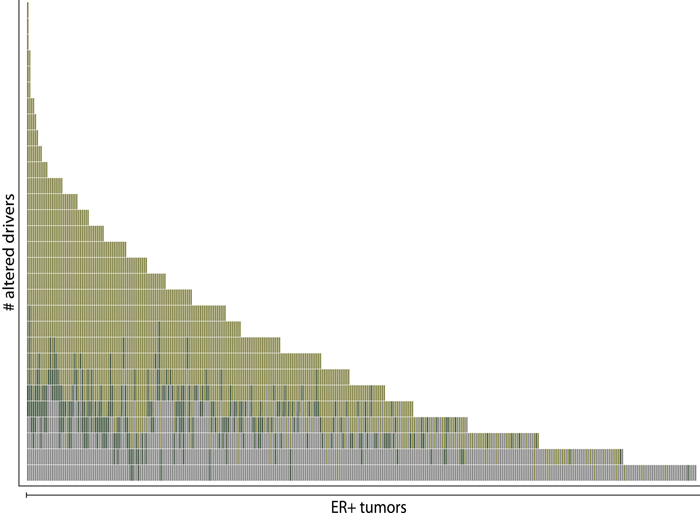
In comparison with a previous study (Stephens et al., 2012, shown in gray), a new computational approach that focuses on somatic copy number mutations increased the number of known driver mutations in breast tumors to a median of five for each tumor. The findings could raise the likelihood of finding actionable targets in individual patients with breast cancer.
For many years, researchers have known that somatic copy number alterations (SCNA’s) — insertions, deletions, duplications, and transpositions of sections of DNA that are not inherited but occur after birth — play important roles in causing many types of cancer. Indeed, most recurrent drivers of epithelial tumors are copy number alterations, with some found in up to 40% of patients with specific tumor types. However, because SCNA’s occur when entire sections of chromosomes become damaged, biologists have had difficulty developing effective methods for distinguishing genes within SCNA’s that actually drive cancer from those genes that might lie near a driver but do not themselves cause disease.
In a new paper published in Cell, researchers in the laboratories of Dana Pe’er (Columbia University Departments of Systems Biology and Biological Sciences) and Jose Silva (Icahn School of Medicine at Mount Sinai) report on a new computational algorithm that promises to dramatically improve researchers’ ability to identify cancer-driving genes within potentially large SCNA’s. The algorithm, called Helios, was used to analyze a combination of genomic data and information generated by functional RNAi screens, enabling them to predict several dozen new SCNA drivers of breast cancer. In follow-up in vitro experimental studies, they tested 12 of these predictions, 10 of which were validated in the laboratory. Their findings nearly double the number of breast cancer drivers, providing many new opportunities towards personalized treatments for breast cancer. Their methodology is general and could also be used to locate disease-causing SCNA’s in other cancer types.
Leading this effort was Felix Sanchez-Garcia, a recent PhD graduate from the Pe’er Lab and a first author on the paper. The story of how this breakthrough came about illuminates how the interdisciplinary research and education that take place at the Department of Systems Biology can address important challenges facing biological and biomedical research.
It started with a dance

Felix Sanchez-Garcia, a recent PhD graduate from the Pe'er Lab, led the development of Helios.
Sanchez-Garcia began his studies at Columbia in 2008 as a master’s student in the Department of Computer Science, specializing in machine learning. During his last semester he began working in Dana Pe’er’s lab, providing statistical validation and support for an earlier algorithm. When he joined the lab, Sanchez-Garcia had little knowledge of the biological sciences, but by working closely with Uri David Akavia, a postdoctoral scientist in the Pe’er Lab, he learned the basics of cancer biology. After completing his master’s he decided to pursue doctoral studies at Columbia, continuing to work in the Pe’er Lab while taking classes in molecular biology, cellular biology, cancer biology, and other related topics. These efforts enabled him to assemble a firm foundation of biological knowledge on which to build his thesis project.
When large cohorts of primary tumor data from projects like the Cancer Genome Atlas (TCGA) became available, the Pe’er Lab began investigating the possibility of studying somatic copy number alterations within the context of cancer research, and found that existing algorithms for identifying cancer-driving SCNA’s had significant limitations. Sanchez-Garcia set out to develop a more robust way for identifying such genomic alterations, working on glioblastoma and ovarian cancer before turning to breast cancer.
A critical moment in the development of the new algorithm occurred not in the laboratory, but on the dance floor. While participating in a recreational swing dancing club at Columbia, Sanchez-Garcia met Ruth Rodriguez Barrueco, a postdoctoral scientist in the Silva Lab who was also working on breast cancer. (At the time, Dr. Silva was based at Columbia University Medical Center.) Silva had already pioneered the use of genome-wide pooled-RNAi screening, which provides valuable functional information about the genome by knocking out each individual gene in a cell line, one at a time. By using computational approaches to analyze RNAi screening data for breast cancer cells, the researchers thought, it might be possible to gain a better understanding of the SCNA’s that drive specific breast cancer subtypes. The work described in the new Cell paper would arise from these conversations.
Predicting SCNA drivers of breast cancer: ISAR and Helios
The method reported in the paper incorporates two algorithms. The first, called Identification of Significantly Altered Regions (ISAR), improves upon previous algorithms for identifying significant SCNA’s, which are expected to harbor a driver gene, by accounting for variations in the local rate at which copy number alterations occur, due to features such as DNA secondary structure and epigenetic alterations. When the researchers applied ISAR to 785 breast cancer samples, they identified 83 significantly amplified SCNA-containing regions, more than doubling the 30 regions previously reported in TCGA. Their findings captured regions identified by previously existing algorithms as well as many novel regions, including several containing known oncogenes. This provided strong evidence that the algorithm was making accurate predictions.
Because the researchers’ ultimate goal was to identify the specific driver genes within these amplified regions that drive breast cancer, Sanchez-Garcia and Pe’er developed a second algorithm, called Helios, which integrates additional information — in this case including point mutation, gene expression, and functional RNAi screening data — into a single candidate driver score. By using machine learning techniques to identify complementary patterns within these diverse data types, Helios iteratively prioritizes genes within significantly altered regions that have the highest probability of being true cancer drivers. As Sanchez-Garcia explains, “Other people used copy number variation to try to narrow things down to a single gene, which is impossible for many regions. Instead, we use copy number to guide interpretation of all the other features.”
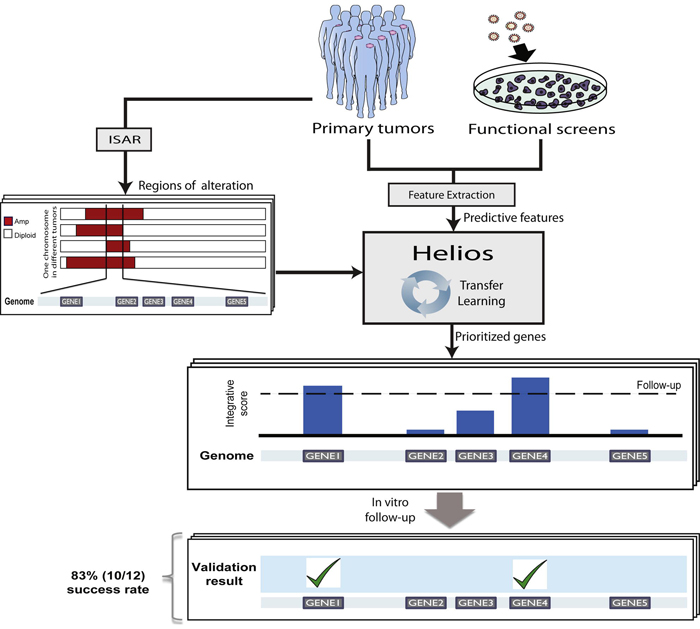
Helios produces an integrated score by combining a set of features derived from primary tumors and genome-wide shRNA screens, and prioritizes genes based on their location in regions of the genome that contain significant copy number alterations. Genes that score above a threshold of significance are then tested experimentally. Helios was highly accurate in predicting cancer driving genes within SCNA's.
Using this integrative approach, Helios was highly accurate in identifying cancer-driving genes. It correctly scored 13 out of 14 (93%) drivers ranked highest within significantly amplified regions. In addition, 10 out of 12 genes (83%) that Helios predicted to be cancer drivers were validated in experimental studies. Reflecting on these results, Dr. Pe’er explains, “This is the first and largest scale systematic validation undertaken for an algorithm of this type, and its accuracy is unprecedented. Its ability to reveal so many new cancer drivers is due to the fact that our hypotheses were generated in an unsupervised way using statistical criteria, rather than by cherry-picking our candidates based on prior biological knowledge. The experimental results show that Helios is a very robust algorithm that can generate biological insights that would be extremely difficult to produce in any other way.”
The results dramatically expand the landscape for investigating drivers of breast cancer, and offer enormous potential for translational breast cancer research. Copy number alterations are known to be much more common than point mutations in patients with breast cancer, which means that this paper’s findings offer the opportunity to identify diagnostic and therapeutic strategies that could improve treatment for large numbers of women with the disease.
Moreover, although used in this paper to study breast cancer, Helios is capable of identifying somatic copy number alterations in many other cancer types in which SCNA’s play a role. It is also designed to be flexible in terms of the biological features it uses as input. “The power of functional screens is significant, but Helios does not require RNAi data,” Garcia-Sanchez explains. “We could use completely different features in our analysis, or investigate diseases such as ovarian, lung, or pancreatic cancer, and it should still work.”
Interdisciplinary training makes the difference
Garcia-Sanchez credits his ability to lead the development of Helios to the combination of computational and biological expertise that contributed to his training and mentorship as a graduate student at Columbia. “Graduate school was challenging,” he recalls, “but taking those biological courses from the beginning of my studies and working in such a collaborative, interdisciplinary environment as Dana’s lab were both very important. To develop the modeling approaches that went into Helios, it was critical to have both a strong understanding of machine learning methods and a deep appreciation of cancer biology and genomics. The modeling choices we made absolutely relied on this. Also, the most exciting research projects will always be the ones that you define yourself, and so having a blend of biological and mathematical knowledge helps you to spot good opportunities for making a difference.”
Dr. Pe’er also acknowledges the Department of Systems Biology and its Center for Multiscale Analysis of Genomic & Cellular Networks (MAGNet) for their commitment to orienting interdisciplinary research around driving biological projects that provide a critical proving ground for computational innovations. “One thing that is unique about this paper is that it wasn’t the case of a student merely applying a computational method to a pre-existing data set. This flips things around, so that the key advance is the innovation that goes into the algorithm, which the experimental lab then validates. Through this process, you also produce useful biological insights. The ability to do this requires an interdisciplinary culture and the right kinds of relationships between computational and experimental scientists.”
Now that his PhD is complete, Sanchez-Garcia is living in Cambridge, England, and is taking time off from academia to work in an industry job in which he can apply his machine learning skills. He hasn’t left biology completely behind, however. He continues to collaborate with the Pe’er and Silva Labs, and is also contributing to research at the Sanger Institute. He remains particularly excited about an upcoming paper that will extend the work described in the Helios paper, remarking, “We’re in the middle of it and there’s still a lot of work to do, but I think if we can get it right, we will be able to identify new therapeutic approaches that could potentially have an enormous benefit to patients.”
— Chris Williams
Related publication
Sanchez-Garcia F, Villagrasa P, Matsui J, Kotliar D, Castro V, Akavia UD, Chen BJ, Saucedo-Cuevas L, Rodriguez Barrueco R, Llobet-Navas D, Silva JM, Pe'er D. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014 Dec 4;159(6):1461-75.