News
Q+A with Dr. Laura Landweber
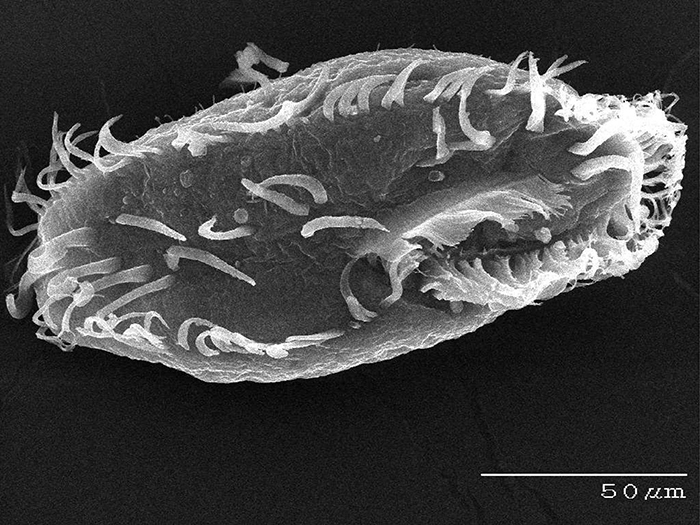
Oxytricha. (Credit: Bob Hammersmith)
Laura Landweber, PhD, loves a challenge. So it’s no surprise that she has built a scientific career unraveling the hows and whys of a unique single-cell organism known for its biological complexity.
An evolutionary biologist whose work sits at the interface of genetics and molecular biology, Dr. Landweber, for nearly 20 years, has focused much of her research on Oxytricha trifallax , a microbial organism that is prevalent in ponds, feeds on algae and has a highly complex genome architecture, making it an attractive, albeit challenging, model organism to study. Compared to humans, with 46 chromosomes containing some 25,000 genes, Oxytricha is known to comprise many thousands of chromosomes, in the ballpark of 16,000 tiny “nanochromosomes”. Yet not only is it complex in sheer numbers of chromosomes but the information carried in those individual chromosomes can be scrambled, like information compression, and the process of development in Oxytricha must descramble this information so that it can be converted into RNA and proteins.
“DNA can be flipped and inverted in Oxytricha and the cellular machinery actually knows how to restore order,” says Dr. Landweber. “Hence, it’s this wonderful paragon for understanding genome integrity and the maintenance and establishment of genome integrity.”
Even more perplexing, in cell division, Oxytricha reproduces asexually when it wants to produce more in number, and it reproduces sexually when it needs to rebuild its genome. It also has the ability to “clean up” its genome, so to speak, eliminating nearly all of the non-coding DNA, or so-called junk DNA. Much of why Oxytricha presents such an intricate genomic landscape remains a mystery, and for Dr. Landweber, the leading expert on this single-celled protist, that wide-open field for potential discovery is what got her hooked.
“We’re learning from Oxytricha a lot about the range of plasticity of genomes and the evolution of genomes, and that our notion of what is a genome doesn’t have to be based on the few model systems that have been studied so far,” she notes. “Most eukaryotes that are studied are animals, but the greater diversity found on our planet is still in the microbial eukaryotic world.”
Dr. Landweber joined Columbia in 2016, after serving on the faculty at Princeton University for over 20 years. The first course she taught at Princeton was rather unconventional, a freshman seminar titled “Jurassic Park: Myth or Reality,” to engage new undergraduates in science subjects from an interdisciplinary standpoint. She says, “It was great fun to discuss the science behind the movie, when both genome sequencing and the study of ancient DNA were new and controversial.”
This spring at Columbia, she will be teaching a graduate/undergraduate seminar on RNA biology, focused on the ancient and modern RNA worlds. The course will begin with a discussion about the origin of life and conclude with the many types of small and large RNAs that flood modern cells.
Early in her tenure at Princeton, Dr. Landweber attended a lecture given by the late David Prescott, PhD, from the University of Colorado-Boulder. Dr. Prescott, at the time, had been studying chromosomes in Oxytricha , and uncovered its unique and complicated genetic makeup that intrigued Dr. Landweber, and set her on the research path she is on today.
Studying this corner of the eukaryotic world provides the Landweber lab with a plethora of research directions. A two-part lab, both experimental and computational, the Landweber group is currently exploring topics of genome rearrangement, and the roles of small and long non-coding RNAs in programming genome rearrangement, as well as genome evolution and molecular evolution. Several of the basic biology discoveries made in Oxytricha have been shown to extend to other organisms.
Dr. Landweber has joint appointments in the Department of Biochemistry and Molecular Biophysics , the Department of Systems Biology and the Department of Biological Sciences . She has been the recipient of a Guggenheim fellowship, an NSF CAREER Award and the Blavatnik Award for Young Scientists. She recently served as president of the Society for Molecular Biology & Evolution.

Laura Landweber, PhD
Oxytricha continues to mystify because of its bizarre genome architecture. You mentioned also that it can “clean up” its genome. What does this mean exactly and what can be gleaned from this?
Oxytricha has the ability to take its genome, which is sort of an informational mess, put it back together and establish these conventional chromosomes. Or, it can establish chromosomes that encode conventional information. It smashes its genome into roughly a quarter million DNA pieces and then rebuilds it. Why does it do that? We of course don’t have the answer yet to that basic question, but one of the motivating factors is to understand why nature allowed a system like this to become so exaggerated and complex. It would be a nice direction to go into in the future to see how the process of Oxytricha’s genome rearrangement can inform us about the landscape of possible genome rearrangements gone awry, like in some cancers, for instance.
What is one theory?
We think it could be runaway evolution. One of the key things we study in Oxytricha is that it has this error-correcting machinery available to it. And, because it can correct errors, it has the ability to buffer the consequences of its mutational burden. And so if mutations happen, they can be erased at the stage of genome assembly during development.
A lot of the pathways that it uses to do this are just variations of pathways that all eukaryotes possess, or nearly all eukaryotes. In that regard, Oxytricha has tweaked some fundamental molecular biology for its benefit to become a really good proofreader, and that sure would be useful as a tool in the future for engineering healthier cells.
Is that possible?
Probably down the road. For now, we’re focused on discovery, the basic science research.
You’ve been investigating Oxytricha for roughly two decades. What continues to intrigue you?
I’m still amazed at how accurately this system can be executed considering how many possible points of failure there are. There’s a quarter of a million DNA pieces that have to be joined correctly and if a few of those are joined incorrectly then the cell may not survive. Like any cell, it might be able to tolerate some error but it’s amazing to me that this process of development that can take just a few days, can bash a genome into a quarter million pieces and put it back together with fidelity, and it does this on the basis of cross checking. It checks its own nascent genome against either mom’s or dad’s reference genome from the previous generation—it has this wonderful cross-talk between generations mediated by RNA molecules.
What’s the most surprising thing, thus far, you’ve learned from your research?
Maybe that [Jean-Baptiste] Lamarck was partly right. Lamarck has a bad reputation for this notion of the transfer of acquired traits. Our work was coming out at the same time as other studies demonstrating that RNA can be a bona fide vehicle for the transmission of acquired characters across generations. Discoveries that we published in 2008 and 2012, as well as results from other labs, demonstrated that RNA can transmit information across generations. And that was pretty exciting. Not that we set out to prove that Lamarck had some merit, but it was becoming vogue again to think that inheritance can depend not only on the DNA genome but on cytoplasmic factors which can include RNA.
Beyond that, we’ve learned that non-coding RNAS inherited from the parents’ cytoplasm, not just messenger RNAs, provide a crucial instructions set to the offspring. We published in Nature in 2008 that long non-coding RNAs can guide genome rearrangement. Then in 2012, we found that a set of millions of smaller RNAs, just 27 nucleotides long, can mark the parts of the genome that it needs to keep, thereby instructing the next generation offspring on how to rebuild their genome. In creating that new genome of 16,000 chromosomes the organism can throw out nearly all of the non-coding DNA, or junk, that it doesn’t need anymore, at least for several asexual generations, and it knows which pieces to keep on the basis of those that are marked by small RNAs, like the post-it notes that mark what stays and what goes. And then the long RNAs that we discovered in 2008 provide the information about the order and orientation to assemble those tiny pieces of DNA.
How do you incorporate systems biology approaches in your research?
There are so many pieces to this Jigsaw puzzle. The Oxytricha genome itself begins at about a billion nucleotides. That gets shattered to roughly a quarter of a million stretches of DNA and the intervening segments are destroyed, so this is systems biology in the sense that we are dealing with problems of large numbers and the cell has to determine, for instance, which of these pieces to keep, which ones to eliminate. And then, there are many millions of 27-nucleotide piRNAs involved in the discrimination of sense and nonsense, and ultimately there are steps that detect and correct error that has crept into this system during post-zygotic development.
We use a lot of computational resources and tools because we’re doing comparative genomics not just across species but even within a single cell. A single cell in Oxytricha has two genomes; And so, to understand the genome architecture of just that one cell, we have to understand and decrypt the genome of both its germline nucleus, which is the archive of all of the information, and the somatic rearranged nucleus, which is the one that has expressed and has captured all the relevant information out of the archive.
Another angle that inspired me to move into this system, besides being awestruck by it, was an analogy to computation. There are parallels in this system to processes of both encryption and decryption, with evolution supplying the encryption of information in DNA, followed by a developmental decryption algorithm that restores the coding capacity of its genome and generates useful chromosomes. Our goal is to reverse engineer how the cell can execute these algorithmic ways of processing its genome. Most genomes don’t require nearly as much processing as Oxytricha’s does.
What is your group working on now?
On the computational front we’re really exploring comparative genomes, on both developmental and evolutionary time-scales. We have a good understanding of the genome architecture and how complex it is in Oxytricha , but from the evolutionary perspective, even if this genome is a Rube Goldberg-like device, the question is how it got that way. So, to understand how this came into being, we have to crawl back on the evolutionary tree and look at antecedents or earlier diverged representatives of life on our planet that are a little bit simpler than Oxytricha . We’re using a lot of the methods that are on the horizon for tools for genome sequencing to probe these other microbial eukaryotes to try to figure out both how and when Oxytricha’s genome became scrambled, and became so highly complex.
-Melanie A. Farmer
Related publications
Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF . RNA-mediated Epigenetic Programming of a Genome-Rearrangement Pathway . Nature. 2008 Jan 10; 451(7175): 153-8.
Fang W, Wang X, Bracht J, Nowacki M, Landweber LF . Piwi-Interacting RNAs Protect DNA Against Loss During Oxytricha Genome Rearrangement . Cell. 2012 Dec 7; 151(6): 1243-1255.