News
Study Sheds Light on Ashkenazi Jewish Genome and Ancestry
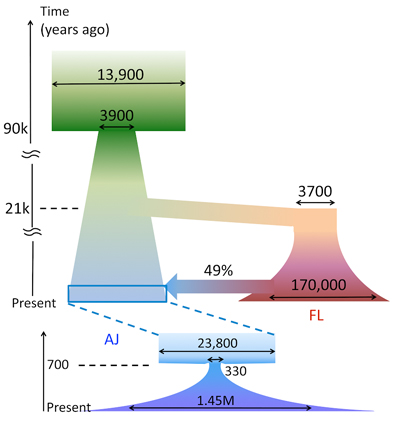
The consortium’s model of Ashkenazi Jewish ancestry suggests that the population’s history was shaped by three critical bottleneck events. The ancestors of both populations underwent a bottleneck sometime between 85,000 and 91,000 years ago, which was likely coincident with an Out-of-Africa event. The founding European population underwent a bottleneck at approximately 21,000 years ago, beginning a period of interbreeding between individuals of European and Middle Eastern ancestry. A severe bottleneck occurred in the Middle Ages, reducing the population to under 350 individuals. The modern-day Ashkenazi community emerged from this group.
An international research consortium led by Associate Professor Itsik Pe’er has produced a new panel of reference genomes that will significantly improve the study of genetic variation in Ashkenazi Jews. Using deep sequencing to analyze the genomes of 128 healthy individuals of Ashkenazi Jewish origin, The Ashkenazi Genome Consortium (TAGC) has just published a resource that will be much more effective than previously available European reference genomes for identifying disease-causing mutations within this historically isolated population. Their study also provides novel insights into the historical origins and ancestry of the Ashkenazi community. A paper describing their study has just been published online in Nature Communications.
The dataset produced by the consortium provides a high-resolution baseline genomic profile of the Ashkenazi Jewish population, which they revealed to be significantly different from that found in non-Jewish Europeans. In the past, clinicians’ only option for identifying disease-causing mutations in Ashkenazi individuals was to compare their genomes to more heterogeneous European reference sets. This new resource accounts for the historical isolation of this population, and so will make genetic screening much more accurate in identifying disease-causing mutations.
In an article that appears on the website of Columbia University’s Fu Foundation School of Engineering and Computer Science, Dr. Pe’er explains:
“Our study is the first full DNA sequence dataset available for Ashkenazi Jewish genomes... With this comprehensive catalog of mutations present in the Ashkenazi Jewish population, we will be able to more effectively map disease genes onto the genome and thus gain a better understanding of common disorders. We see this study serving as a vehicle for personalized medicine and a model for researchers working with other populations.”
In addition to offering an important resource for such future translational and clinical research, the paper’s findings also provide new insights that have implications for the much debated question of how European and Ashkenazi Jewish populations emerged historically.
By analyzing the proliferation of long nucleotide sequences that are identical in the unrelated individuals in their sample, the researchers determined that a population bottleneck of approximately just 350 Ashkenazi Jewish individuals occurred in central Europe about 700 years ago, followed by an exponentially rapid population increase. The findings suggest that the ancestry of all present-day Ashkenazi Jews can be traced back to this small population.
Moreover, by comparing the genomes of Ashkenazi Jews with those of Flemish origin, the researchers found strong evidence that the ancestry of the modern-day Ashkenazi can be traced to a fairly even mixture of European and Middle Eastern descent. Cross-breeding appears to have occurred at approximately the same time as the Ashkenazi bottleneck, suggesting that when Jewish migrants arrived in Europe from the Levant they mixed with the local population.
The consortium’s findings also indicate that modern European ancestry can be traced to migration from the Middle East following the last great ice age, approximately 12,000-25,000 years ago. Others have argued that Europe was populated by an earlier migration from Asia more than 40,000 years ago, though this study provides strong evidence against that hypothesis.
The whole genome sequence data produced by this study are available at the European Genome-Phenome Archive. Dr. Pe’er anticipates that it will become a resource for other scientists interested in population genetics and personalized medicine.
— Chris Williams
Related publications
Carmi S, Hui KY, Kochav E, Liu X, Xue J, Grady F, Guha S, Upadhyay K, Ben-Avraham D, Mukherjee S, Bowen BM, Thomas T, Vijai J, Cruts M, Froyen G, Lambrechts D, Plaisance S, Van Broeckhoven C, Van Damme P, Van Marck H, Barzilai N, Darvasi A, Offit K, Bressman S, Ozelius LJ, Peter I, Cho JH, Ostrer H, Atzmon G, Clark LN, Lencz T, Pe'er I. Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nat Commun. 2014 Sep 9;5:4835.
Visit The Ashkenazi Genome Consortium for additional coverage.