News
Synergy between Two Genes Drives Aggressive Prostate Cancer
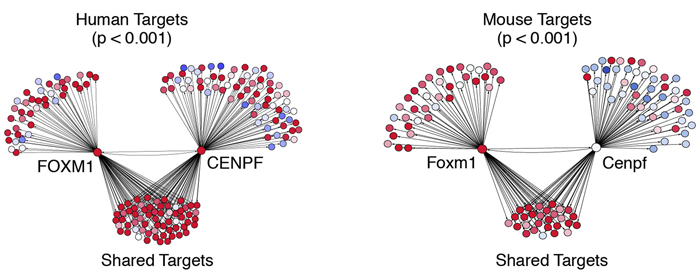
Computational synergy analysis depicting FOXM1 and CENPF regulons from the human (left) and mouse (right) interactomes showing shared and nonshared targets. Red corresponds to overexpressed targets and blue to underexpressed targets.
Two genes work together to drive the most lethal forms of prostate cancer, according to new research by investigators in the Columbia University Department of Systems Biology. These findings could lead to a diagnostic test for identifying those tumors likely to become aggressive and to the development of novel combination therapy for the disease.
The two genes—FOXM1 and CENPF—had been previously implicated in cancer, but none of the prior studies suggested that they might work synergistically to cause the most aggressive form of prostate cancer. The study was published today in the online issue of Cancer Cell.
“Individually, neither gene is significant in terms of its contribution to prostate cancer,” said co-senior author Andrea Califano, the Clyde and Helen Wu Professor of Chemical Biology in Biomedical Informatics and Chair of the Department of Systems Biology. “But when both genes are turned on, they work together synergistically to activate pathways associated with the most aggressive form of the disease.”
Co-principal investigator Andrea Califano discusses the new study.
“Ultimately, we expect this finding to allow doctors to identify patients with the most aggressive prostate cancer so that they can get the most effective treatments,” said co-senior author Cory Abate-Shen, the Michael and Stella Chernow Professor of Urologic Sciences and also a member of the Department of Systems Biology. “Having biomarkers that predict which patients will respond to specific drugs will hopefully provide a more personalized way to treat cancer.”
Scientists widely recognize that cancer is characterized by multiple genetic changes. “However, distinguishing the handful of genes that are driving the cancer from the many genes whose altered expression does not contribute directly to the cancer has proven to be a daunting task,” said Dr. Califano. “It becomes even more difficult when genes work together synergistically, because they must be analyzed in pairs rather than one by one. For instance, the approximately 1,000 genes that have been linked to cancer can be combined into about 500,000 gene pairs, each of which may represent a synergistic tumor driver. This is an enormous number that defies our best statistical tools and requires sophisticated systems biology approaches.”
“Prostate cancer is particularly challenging because it has such a wide variety of clinical presentations, with relatively few shared genetic mutations,” said Dr. Abate Shen.
Thus, to find the key genes that drive prostate cancer, the CUMC team devised a novel experimental approach in which they used computational approaches to compare the gene regulatory networks that drive prostate cancer in humans with those in a genetically engineered mouse model of the disease.
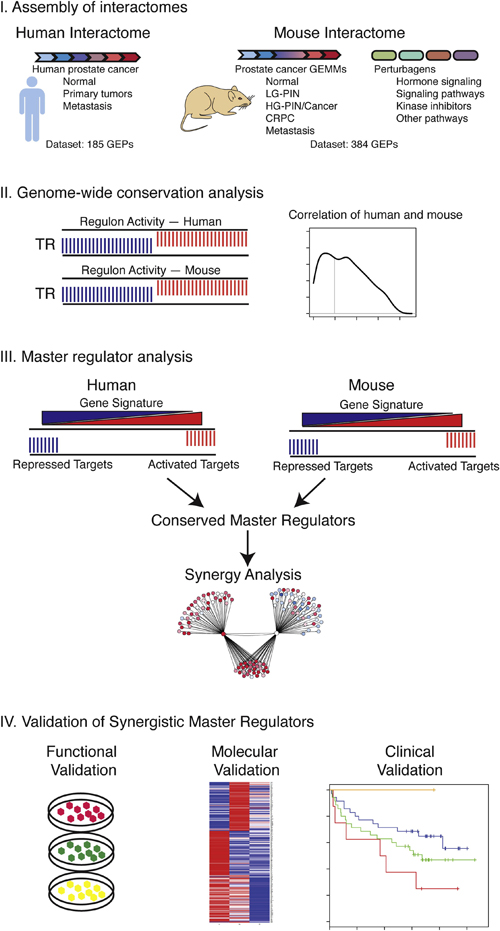
Strategy for Genome-wide Cross-Species Analyses of Prostate Cancer. Schematic representation of the overall strategy. Step I: assembly of human and mouse prostate cancer interactomes. Step II: genome-wide computational analysis of conservation of transcriptional regulon activity in the mouse and human prostate cancer interactomes. Step III: master regulator analysis for identification of conserved master regulators and prediction of synergy. Step IV: validation of candidate master regulators using functional, molecular, and clinical analyses.
In many cancer studies, researchers rely on mouse models to identify genes that are expressed in disease. “However, inherent differences between species often make it difficult to extrapolate findings in mice to humans,” said Dr. Abate-Shen.
“By tracing the regulatory logic of these tumors in both species,” said Dr. Califano, “we were able to identify identical driver genes of malignant prostate cancer and to discover that they don’t work as individual drivers but rather together, as a synergistic driver pair.”
Using the high-performance computing cluster housed in the Department of Systems Biology, the analysis identified FOXM1 and CENPF as a synergistic driver pair in aggressive prostate cancer in both mice and humans, as these regulators jointly control genetic programs associated with the most prominent tumor hallmarks in both species. Individually, the aberrant expression of these genes does not activate these programs. When acting together, however, the two genes can wreak havoc in the cancer cell and turn it into a very aggressive tumor.
To validate the roles of FOXM1 and CENPF, the researchers silenced the expression of the genes in four human prostate cancer cell lines, first individually and then together. Silencing either gene alone had only a modest effect on the ability of the cells to form tumors. However, co-silencing both genes at once completely stopped the growth of tumors in a mouse. This observation is consistent with a synergistic interaction, where the joint effect of both genes is much greater than the sum of their individual effects.
The researchers then analyzed prostate cancers from a group of more than 900 patients who had undergone prostate removal surgery. This analysis showed a striking correlation between the co-expression of FOXM1 and CENPF and the poorest disease outcome. In sharp contrast, expression of either gene alone did not correlate with aggressive disease. In addition, tumors in which neither gene was aberrantly expressed had the best prognosis.
The researchers also showed that silencing the two genes inactivated the PI3-kinase and MAP kinase signaling pathways, both of which are known to be hallmarks of aggressive prostate cancers. “This adds additional strength to the possibility that combined therapeutic targeting of both FOXM1 and CENPF may be effective in arresting the human disease,” says Dr. Abate-Shen.
“This is just a first step toward a deeper understanding of the genetics of cancer,” said Michael Shen, also a member of the Department of Systems Biology and a contributor to the study. “The tools and approaches developed in this study may have broad utility in studying prostate cancer; cross-species computational analyses also could be used to identify the causes of other cancers, as well as that of other complex diseases.”
Article adapted with permission of Columbia University Medical Center.
Reference:
Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, Califano A, Abate-Shen C. Cross-Species Regulatory Network Analysis Identifies a Synergistic Interaction between FOXM1 and CENPF that Drives Prostate Cancer Malignancy. Cancer Cell. 2014 May 12;25(5):638-51.