News
Improving Single-Cell RNA-Seq at the Columbia Genome Center
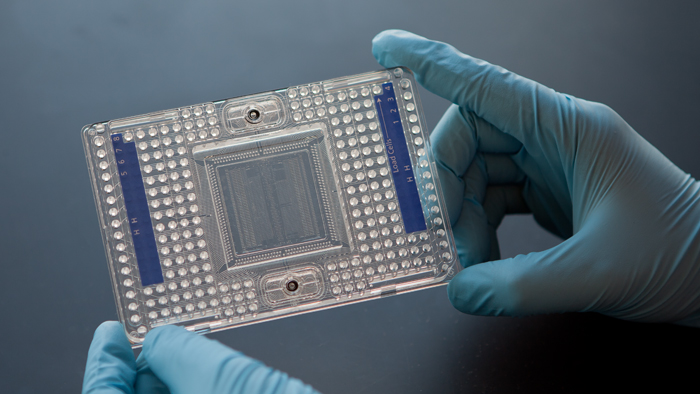
At the core of the Fluidigm C1 Single-Cell Auto Prep System is a 96-well plate containing microfluidics. After individual cells are isolated in their own wells, the device amplifies their cDNA for genome-wide gene expression profiling. Scientists at the Columbia Genome Center are developing methods for addressing the technical and analytical challenges of single-cell RNA sequencing, and have begun generating some exciting data.
Since the invention of the first microscope, a procession of new technologies has enabled scientists to study individual cells at increasingly fine levels of detail. The last two years have witnessed an important next stage in this evolution, with the arrival of the first devices for genetically profiling single cells on a genome-wide scale.
The first commercial product in this field is the Fluidigm C1 Single-Cell Auto Prep System, which uses microfluidics to isolate single cells and offers the ability to generate gene expression profiles for up to 96 cells at a time. But because of the novelty of the technology and the inherent difficulties of working with single cells, it has presented a number of technical challenges for researchers interested in exploring biology at this level.
Now, scientists at the JP Sulzberger Columbia Genome Center led by Assistant Professors Peter Sims and Yufeng Shen have developed an experimental and computational pipeline that optimizes the C1’s capabilities. And even as they work to solve some of the challenges that are inherent to single-cell research, their approach has begun generating some exciting data for studying genetics in a variety of cell types.
Addressing the problem of cellular heterogeneity
The Fluidigm C1 and other emerging methods for single-cell sequencing have attracted so much interest in the research community because they address a limitation of population-level or bulk sequencing. By design, the data that are output from a typical whole genome, exome, or RNA sequencing experiment are based on the averages of millions of reads taken across large populations of cells. Although these approaches have become invaluable to biological and biomedical research, they are not intended to capture the inherent variability among individual cells, a heterogeneity that research is showing to have clear importance in areas such as development, disease progression, and how diseased cells respond to drugs.
In basic biological research, it is common to define a population of cells based on a limited number of molecular markers. Nevertheless, when subjected to specific perturbations, individual cells within that population can sometimes behave differently, suggesting the existence of subpopulations that are distinct in other ways. For example, in a pool of stem cells that appear to be identical, some might not mature as quickly as others, or individual cells might differentiate into different types of tissue under the same stimulus. Heterogeneity is also seen as an important problem in cancer, as cells in the same tumor can respond in varying degrees to the same drug treatment. In these and many other situations, bulk genome profiling methods do not have the resolution to distinguish why one cell behaves in one way, and another behaves differently.
By isolating individual cells and amplifying their genetic information, the Fluidigm C1 makes it possible to observe and compare gene expression in single cells, offering a high-resolution method for studying the origins of this kind of heterogeneity.
The Columbia Genome Center’s experimental pipeline
Currently, the Fluidigm C1 is designed to facilitate single-cell RNA sequencing, which provides an unbiased picture of the complete set of RNA molecules present in the cell (also called the cell's transcriptome). The Columbia Genome Center has already used the Fluidigm C1 to perform single-cell RNA-Seq on several different cell types, including T cells, dendritic cells, tumor cells, and stem cells. In doing so they have developed a pipeline that addresses the technical challenges of sequencing single cells, and can account for differences among various cell types that can influence how the machine is used.
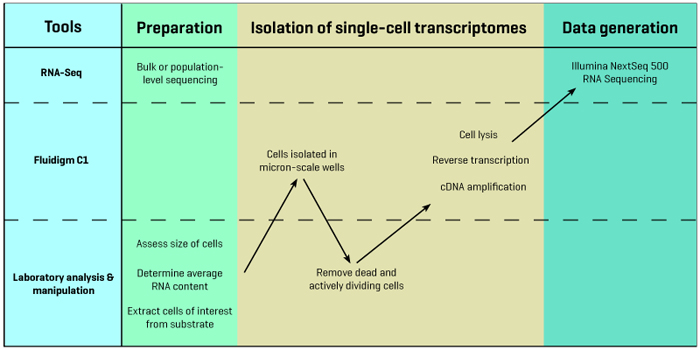
The Columbia Genome Center adds several steps to the basic Fluidigm C1 pipeline in order to optimize the device's capabilities. These steps make use of a microscope for two-color fluorescence imaging maintained in the Sims Lab as well as next-generation sequencing infrastructure in the Columbia Genome Center. Bulk or population-level RNA sequencing is performed early in the pipeline as a way of verifying the accuracy of the single-cell sequencing results.
The first step in the Genome Center’s pipeline is to determine the average size of the cells that will be studied. This is important because Fluidigm offers a variety of chips with different microfluidics configurations, depending on the size of the cells to be sequenced. After a collaborator interested in pursuing single-cell research grows or isolates a sample population of cells, researchers in the Genome Center use a high-resolution light microscope to image the cells and carefully determine their size distribution.
Before placing the cells in the C1, they also conduct experiments to assess the average amount of RNA found in each cell. This is important for calculating how much reagent to administer during sequencing. The reagents include standard oligonucleotides at known concentrations that are used for normalization, and so if there is not a large amount of RNA in each cell, adding too much reagent can obscure the gene expression signal and lead to inaccurate results.
Before beginning the actual single-cell sequencing experiment, the Genome Center also performs bulk or population-level sequencing on the entire test sample, providing a baseline average readout of gene expression levels. This information becomes an important reference following single-cell sequencing, as the average overall single-cell expression levels should approximate the expression levels seen in the bulk sample.
Following these preliminary experiments, cells of interest are isolated from a suspension or dissociated from their substrate (for example, from a culture dish or a solid tissue) and sorted using markers. The sample is then loaded into Fluidigm’s chip, which uses microfluidics to distribute individual cells into discrete micron-scale chambers.

On the left, Peter Sims operates a sophisticated microscope used to physically inspect cells before sequencing. The image on the right shows a single cell that has been isolated within the Fluidigm chip's microfluidic canals.
At this point, the researchers remove the plate from the C1 and use two-color fluorescence imaging to distinguish live cells from dead cells, wells that contain more than one cell, and cells that are actively dividing. Identifying and then disregarding these cells helps to avoid the unnecessary costs of sequencing cells that will not provide useful information. Once this step is complete, an average of 50 viable cells typically remain for single-cell sequencing.
Following this sorting step the researchers return the plate to the C1, which lyses the individual cells, reverse transcribes their cDNA, and then amplifies the cDNA. This step yields one pre-amplified cDNA library for each cell that was processed in the C1. These libraries are then prepared for sequencing using the Columbia Genome Center’s Illumina NextSeq 500 sequencer. Once sequencing begins, raw data for the cells are typically available within just 24 hours.
Ongoing challenges of single-cell sequencing
There are a number of important differences between single-cell sequencing and bulk sequencing. For one, the amount of RNA in a single cell is very low, so the corresponding cDNA must be amplified extensively. This can add statistical noise to the signal due to inherent biases in the necessary chemistry.
Moreover, in traditional genomic studies, analysis often involves comparing two different but clearly defined samples (for example, diseased cells vs. healthy cells) to identify genetic differences associated with a phenotype. With single cell sequencing, the problem is flipped on its head. Here, the key question is to find out what even constitutes a subpopulation of cells and the criteria for doing so are unknown. This presents the challenge of not only identifying genomic variants between cells, but also determining which variants are essential in defining each subpopulation. Analyzing the data therefore requires a variety of new computational approaches that are an active field of research in Yufeng Shen’s lab.
Single cell profiling can also sometimes be misleading because RNA expression can fluctuate wildly. RNA typically has a much shorter lifetime than a protein and can be born and die within a matter of minutes, translating many copies of a protein very quickly. Once this burst of activity is complete, however, gene expression can shut down, even though the proteins that resulted from that burst are still present at high concentrations in the cell. Therefore, although the C1 provides an exquisite picture of gene transcription, it doesn’t necessarily correlate to what signaling networks are active in the cell at that time point.
For these and other reasons, Dr. Sims cautions, “Single-cell sequencing is not like bulk sequencing at all. It’s not a well-established technology. We’re really at the frontier here and are going to encounter situations where the technology and the pipeline don’t work well. Fortunately, though, we’ve already made a lot of progress in addressing these problems and have gotten some interesting findings.”
Building on early successes
Despite the unique challenges that single-cell sequencing poses, the pipeline developed at the Columbia Genome Center has already produced a number of encouraging results. Sims predicts that the next year will be a big one for single-cell research. He says that his lab has generated some interesting data on glioblastoma, and at least one early adopter of the technology has conducted multiple, iterative experiments to generate a large data set that is likely to lead to a publication.

The Columbia Genome Center operates the Fluidigm C1 in tandem with the Illumina NextSeq 500. Once the C1 has produced a collection of single-cell cDNA libraries, the NextSeq can generate sequencing data within 24 hours.
At the same time, he says, “The C1 is not at the point yet where it will be an everyday tool that anyone can use. To be successful it requires a pipeline such as the one we’ve developed at the Genome Center, and a lab needs to make a big commitment when they decide to go in this direction with their research. At the same time, though, it’s been fascinating to see some of the early results, since they’re revealing a layer of biological complexity that we’ve never had a way to study before.”
Providing such state-of-the-art tools for the Columbia University research community is an important part of the mission of the JP Sulzberger Columbia Genome Center. “We are building an advanced sequencing infrastructure from which all researchers at Columbia can benefit,” says Olivier Couronne, executive director of the Center. “This is very much a shared vision across the institution, and in developing our single-cell sequencing pipeline we’ve worked closely with the Herbert Irving Comprehensive Cancer Center, the Department of Systems Biology, the Columbia Stem Cell Initiative, and the Department of Pathology, as well as with the Helmsley Charitable Trust, which provided crucial support. We see the development of such partnerships as particularly important because this kind of cutting-edge technology shows enormous potential to have a clear impact on genomic medicine as well as in the clinic, and to shorten the cycle time for discovery.”
— Chris Williams