News
Columbia Investigators Awarded New NCI Physical Sciences in Oncology Center
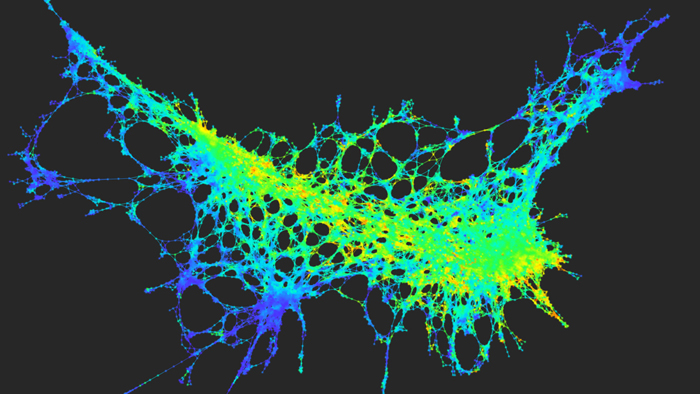
The Columbia University Center for Topology of Cancer Evolution and Heterogeneity will combine mathematical approaches from topological data analysis with new single-cell experimental technologies to study cellular diversity in solid tumors. Image courtesy of Raul Rabadan.
The National Cancer Institute’s Physical Sciences in Oncology program has announced the creation of a new center for research and education based at Columbia University. The Center for Topology of Cancer Evolution and Heterogeneity will develop and utilize innovative mathematical and experimental techniques to explore how genetic diversity emerges in the cells that make up solid tumors. In this way it will address a key challenge facing cancer research in the age of precision medicine — how to identify the clonal variants within a tumor that are responsible for its growth, spread, and resistance to therapy. Ultimately, the strategies the Center develops could be used to identify more effective biomarkers of disease and new therapeutic strategies.
Co-directing the Center are Raul Rabadan, associate professor in the Departments of Systems Biology and Biomedical Informatics, and Antonio Iavarone, professor of pathology and cell biology and neurology, and an investigator in the Columbia University Medical Center’s Institute for Cancer Genetics. The multidisciplinary research team also includes 8 additional investigators based at Columbia, Stanford University, New York University, and the University of Texas at Austin. The researchers will collaborate closely, bringing together expertise in cancer genomics, the genetics of brain tumors, developmental biology, single-cell genomics, and machine learning, as well as a mathematical field called topological data analysis that recent research has shown to hold potential applications for the study of cancer. Other participating scientists include Michael Shen, Peter Sims, Anna Lasorella, Chris Wiggins, Gunnar Carlsson (Stanford), Andrew Blumberg (Texas), Bud Mishra (NYU), and Hossein Khiabanian.
The NCI’s Physical Sciences in Oncology Network was started in 2009 in recognition of the fact that perspectives based in mathematics, physics, chemistry, and engineering all have roles to play in cancer research, and so education and outreach activities that build bridges between communities are an important part of its mission. In addition to conducting research, then, the new Center will create the New York Metropolitan Area Discussion Group in Mathematics and Oncology, which will promote dialogue between physical scientists and mathematicians on the one side, and cancer biologists on the other, in order to facilitate the development of new breakthroughs. Open to all interested scientists, the Discussion Group will be coordinated by investigators at Columbia University Medical Center, NYU’s Courant Institute of Mathematical Science, and the Institute for Advanced Study.
The new Center will also support the cancer research community by providing opportunities for quantitative investigators to receive education in cancer biology and become embedded in biology laboratories; distribute startup grants for interdisciplinary, collaborative research; and organize an annual international conference that will bring together the larger network of investigators working at the intersection of the physical sciences and cancer research.
Tracking genetic diversity in tumor cells
The key scientific issue driving research at the new center at Columbia is an emerging awareness that genetic diversity among the cells that make up solid tumors is a critical factor in determining how cancer grows. Recent research has shown, for example, that gene expression varies in different regions in a single tumor, suggesting variability in how different cells function. A growing body of research has also revealed that the most common mutations in a tumor are not necessarily the ones that drive its growth.
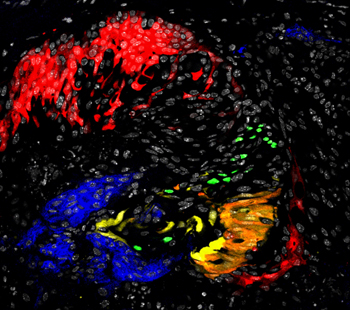
Tracing clonal lineages with fluorescent markers. Image by Flaminia Talos, Michael Shen Lab.
Over the last several years, genome sequencing has helped researchers to uncover genetic variations that distinguish cancer cells from normal cells. However, conventional bulk genome sequencing methods, which find statistical averages of genome sequences across the entire collection of cells in a tumor, are currently limited in their ability to capture the heterogeneity seen in genetically distinct clones. This has made the search for targeted therapies very slow, and has made it difficult to identify more effective strategies for preventing metastasis and drug resistance, which can result from clonal genetic variation.
The Center will approach this problem by developing new interdisciplinary methods for understanding the evolutionary processes at play during tumor development; that is, how genetic diversity emerges from a single cancer-initiating cell. Focusing on prostate cancer and glioblastoma as sample systems, it will develop widely applicable pipelines for modeling tumor evolution, dissecting clonal heterogeneity, and achieving insights into the genetic mechanisms that lead to drug resistance during cancer treatment. These efforts, the Center’s researchers anticipate, should ultimately provide the scientific community with widely applicable models and methods for understanding how individual tumors develop, information that could lead to new biomarkers and treatment strategies.
Working toward this goal, the Center will make use of two key, emerging experimental technologies. One is the organoid culture, in which three-dimensional buds of organs are grown in the laboratory, providing an experimental system that more closely replicates what happens in the body than traditional in vitro cultures. As Center investigator Michael Shen has shown, these can be combined with fluorescent tagging methods that make it possible to track how specific cellular clones divide and evolve during tumor development. A second key technology is high-throughput single-cell sequencing. New methods currently under development in the laboratory of Peter Sims make it possible to quickly generate readouts of the genomes and gene products of large numbers of individual cells. By combining these experimental methods, the Center aims to provide a high-resolution representation of genetic differences among the cells that make up a tumor, making it possible to distinguish how clonal subpopulations evolve.
Topology and cancer data
At the same time that experimental technologies have improved biologists’ ability to reveal clonal heterogeneity, however, they have also created a need for new mathematical approaches for interpreting the large, high-dimensional data sets they generate. In recent work, Raul Rabadan, Gunnar Carlsson, Andrew Blumberg, and other participating scientists have been developing new applications of a mathematical field called topological data analysis (TDA), which they have already shown to provide useful strategies for handling such data. TDA is an advanced method based in the mathematical field of topology that can characterize the relationships and structures underlying large collections of discrete measurements.

The new Center will be led by Raul Rabadan and Antonio Iavarone.
The new Center will extend this work to develop and apply new mathematical approaches for interpreting single-cell data. Using TDA, they will create a mathematical structure for identifying distinct cell populations, and determine their association with the progression of primary solid tumors. Using another mathematical concept called moduli spaces, they will also define methods for comparing “trees” representing clonal evolution from multiple clones. This ability should make it possible to extract information that can be used to provide clinical insights into tumor progression. Finally, they will design algorithms to disentangle relationships among mutational events in tumor evolution, with the goal of distinguishing mutations that truly drive cancer progression from those that are merely associated with it.
By combining experimental and quantitative approaches in this way, the Columbia University Center for Topology of Cancer Evolution and Heterogeneity will deliver validated methods for inferring clonal evolution, new single-cell genomic protocols for uncovering clonal heterogeneity, and experimentally validated machine learning approaches for predicting drug sensitivities. “Our ultimate goal,” Dr. Rabadan says, “will be to provide the cancer research community with a framework for unraveling complexity in solid tumors, with the long-term aim of improving diagnosis and treatment.”