News
Picture This: Integrating Single-Cell Sequencing with Live Cell Imaging
New NIH Grant to Accelerate Commercialization of Single-Cell Analysis Platform
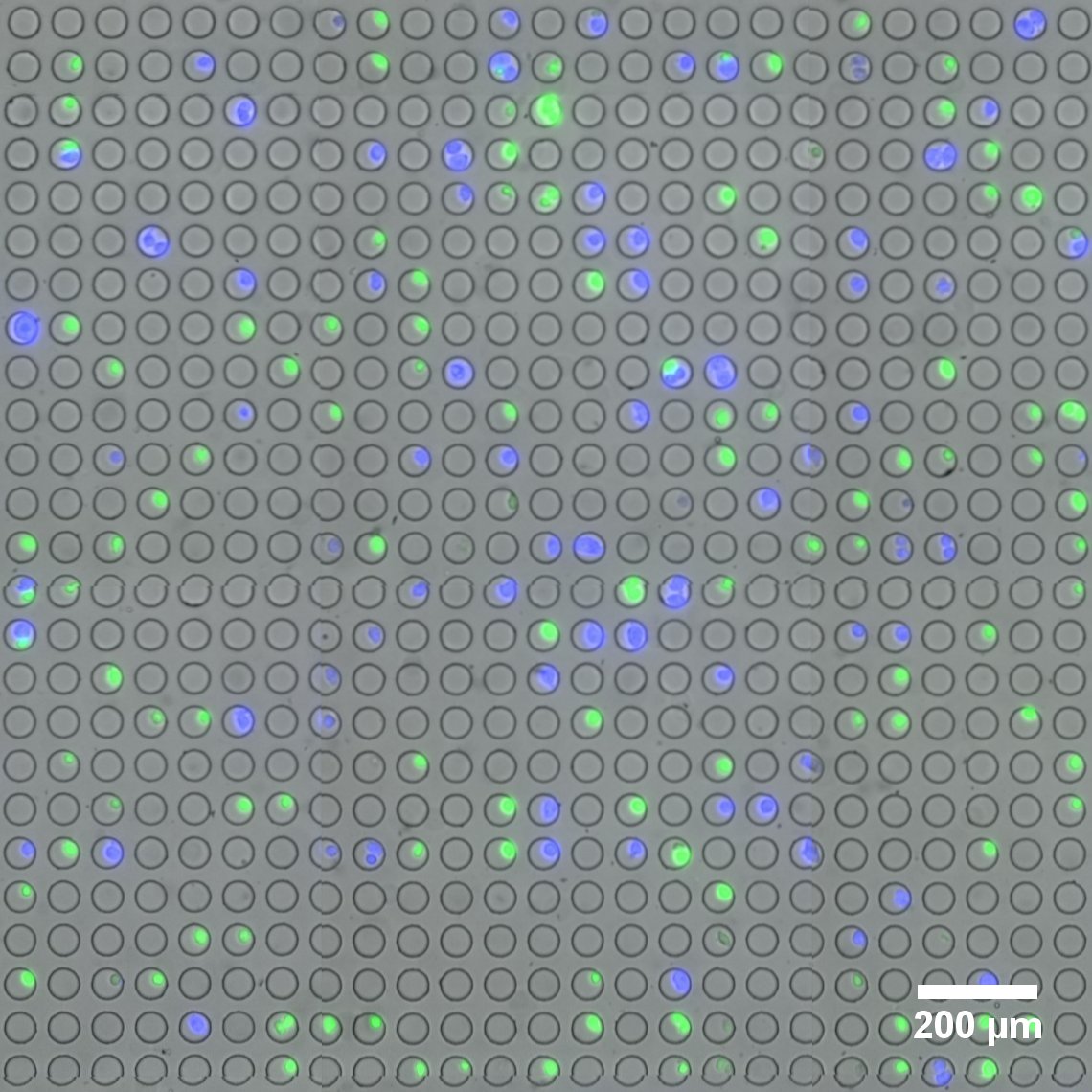
Microwell array flow cell device loaded with fluorescently-labeled live cells. (Image provided by the Sims Lab.)
The field of single-cell RNA sequencing is moving at a fast clip. Adding to its rapid advance is a novel platform for linking optical imaging with high-throughput single-cell sequencing devised by researchers in the Sims Lab at Columbia’s Department of Systems Biology.
The new technology, developed by Peter Sims, PhD , assistant professor of systems biology, and postdoctoral research scientist, Jinzhou Yuan, PhD , enables live cell imaging and RNA sequencing simultaneously of the same individual cell on a large scale and at low cost. Jointly awarded a $1.5 million grant funded by the National Institutes of Health’s SBIR program, the Sims Lab and Cell Microsystems are collaborating to build and test the device, with the goal of bringing to market a fully-integrated system capable of imaging thousands of single cells and preparing them for genomic analysis.
The proposed system will integrate Cell Microsystems’ proprietary CellRaft Technology with the Sims Lab’s novel approaches to tracking single cells with so-called optical barcodes.
In single-cell RNA sequencing, “State-of-the-art technology now allows us to routinely process thousands of individual cells and obtain their genome-wide mRNA expression profiles,” notes Dr. Yuan. “However, cells that share similar expression profiles at the mRNA level may be distinct from each other based on features obtainable by optical microscopy, such as morphology, motility, and fluorescent labels. A more comprehensive description of each cell obtained by both imaging and sequencing may help us further refine cell type and state.”
“We’re leveraging a 400-year-old method—microscopic imaging—and refining it to link nicely with high-throughput sequencing,” says Dr. Sims. “Imaging cells is key. There are a lot of features of a cell you can’t infer without images, such as shape, size, and behavior.”
Drs. Sims and Yuan devised their optical barcoding method, called SCOPE-Seq, to be implemented in a microwell array-based platform, which analyzes thousands of individual cells in parallel, enabling it to detect thousands of genes per cell, maintain high expression profile purity, and link optical phenotypes and expression profile of the same cell with high accuracy. In future work, the SCOPE-Seq technique could be applied to study the molecular mechanism of drug resistance or to associate dynamic protein expression patterns with genome-wide mRNA expression profile of individual cells.
The Sims Lab is an early contributor to the emerging, fast-growing field of automated single-cell RNA sequencing, which has made it possible to analyze tens of thousands of cells but at the same time obtain characteristics and genomics data from each individual cell. This technique is also making it possible to discover new cell types. The Sims group applies cutting-edge microscopy, next-generation sequencing, and microfabrication to enable unbiased, genome-wide measurements in wide-ranging biological systems.
Under this new NIH grant, Cell Microsystems and the Sims Lab will work closely together to build a user-friendly version of the SCOPE-Seq system coupled with Cell Microsystems’ cost-effective method to isolate and recover single cells for direct analysis; ultimately turning their platform into a single-button solution. Additionally, the new optical barcoding approach is contributing to the Human Cell Atlas Project, a global effort to identify and characterize each individual cell type of the human body, with the support of the Chan Zuckerberg Initiative.
Says Sims, “We are excited to work towards commercializing this new platform, which will enable researchers to obtain more comprehensive pictures of individual cells with the scalability for high-throughput applications.”
By Melanie A. Farmer